Cas9 specifies functional viral targets during CRISPR-Cas adaptation
- PMID: 25707807
- PMCID: PMC4385744
- DOI: 10.1038/nature14245
Cas9 specifies functional viral targets during CRISPR-Cas adaptation
Abstract
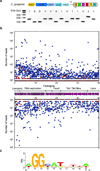
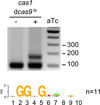
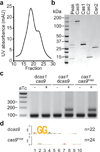
Clustered regularly interspaced short palindromic repeat (CRISPR) loci and their associated (Cas) proteins provide adaptive immunity against viral infection in prokaryotes. Upon infection, short phage sequences known as spacers integrate between CRISPR repeats and are transcribed into small RNA molecules that guide the Cas9 nuclease to the viral targets (protospacers). Streptococcus pyogenes Cas9 cleavage of the viral genome requires the presence of a 5'-NGG-3' protospacer adjacent motif (PAM) sequence immediately downstream of the viral target. It is not known whether and how viral sequences flanked by the correct PAM are chosen as new spacers. Here we show that Cas9 selects functional spacers by recognizing their PAM during spacer acquisition. The replacement of cas9 with alleles that lack the PAM recognition motif or recognize an NGGNG PAM eliminated or changed PAM specificity during spacer acquisition, respectively. Cas9 associates with other proteins of the acquisition machinery (Cas1, Cas2 and Csn2), presumably to provide PAM-specificity to this process. These results establish a new function for Cas9 in the genesis of prokaryotic immunological memory.
Conflict of interest statement
The authors have no conflicting financial interests.
Figures






Comment in
-
Microbiology: How bacteria get spacers from invaders.Nature. 2015 Mar 12;519(7542):166-7. doi: 10.1038/nature14204. Epub 2015 Feb 18. Nature. 2015. PMID: 25707799 No abstract available.
References
Methods references
-
- Duplessis M, Moineau S. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 2001;41:325–336. - PubMed
-
- Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

