Mechanisms of mRNA frame maintenance and its subversion during translation of the genetic code
- PMID: 25708857
- PMCID: PMC4458409
- DOI: 10.1016/j.biochi.2015.02.007
Mechanisms of mRNA frame maintenance and its subversion during translation of the genetic code
Abstract
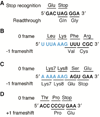
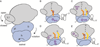
Important viral and cellular gene products are regulated by stop codon readthrough and mRNA frameshifting, processes whereby the ribosome detours from the reading frame defined by three nucleotide codons after initiation of translation. In the last few years, rapid progress has been made in mechanistically characterizing both processes and also revealing that trans-acting factors play important regulatory roles in frameshifting. Here, we review recent biophysical studies that bring new molecular insights to stop codon readthrough and frameshifting. Lastly, we consider whether there may be common mechanistic themes in -1 and +1 frameshifting based on recent X-ray crystal structures of +1 frameshift-prone tRNAs bound to the ribosome.
Keywords: Frameshifting; Protein synthesis; RNA structure; Ribosome; Translocation.
Copyright © 2015 Elsevier B.V. and Société française de biochimie et biologie Moléculaire (SFBBM). All rights reserved.
Figures


Similar articles
-
Dynamic pathways of -1 translational frameshifting.Nature. 2014 Aug 21;512(7514):328-32. doi: 10.1038/nature13428. Epub 2014 Jun 11. Nature. 2014. PMID: 24919156 Free PMC article.
-
Mechanism of tRNA-mediated +1 ribosomal frameshifting.Proc Natl Acad Sci U S A. 2018 Oct 30;115(44):11226-11231. doi: 10.1073/pnas.1809319115. Epub 2018 Sep 27. Proc Natl Acad Sci U S A. 2018. PMID: 30262649 Free PMC article.
-
A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting.Nature. 2006 May 11;441(7090):244-7. doi: 10.1038/nature04735. Nature. 2006. PMID: 16688178 Free PMC article.
-
The many faces of ribosome translocation along the mRNA: reading frame maintenance, ribosome frameshifting and translational bypassing.Biol Chem. 2023 Apr 21;404(8-9):755-767. doi: 10.1515/hsz-2023-0142. Print 2023 Jul 26. Biol Chem. 2023. PMID: 37077160 Review.
-
Changed in translation: mRNA recoding by -1 programmed ribosomal frameshifting.Trends Biochem Sci. 2015 May;40(5):265-74. doi: 10.1016/j.tibs.2015.03.006. Epub 2015 Apr 4. Trends Biochem Sci. 2015. PMID: 25850333 Free PMC article. Review.
Cited by
-
Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.Elife. 2020 Oct 5;9:e51898. doi: 10.7554/eLife.51898. Elife. 2020. PMID: 33016876 Free PMC article.
-
Ablation of Programmed -1 Ribosomal Frameshifting in Venezuelan Equine Encephalitis Virus Results in Attenuated Neuropathogenicity.J Virol. 2017 Jan 18;91(3):e01766-16. doi: 10.1128/JVI.01766-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27852852 Free PMC article.
-
Model of the pathway of -1 frameshifting: Long pausing.Biochem Biophys Rep. 2016 Jan 29;5:408-424. doi: 10.1016/j.bbrep.2016.01.017. eCollection 2016 Mar. Biochem Biophys Rep. 2016. PMID: 28955849 Free PMC article.
-
Model of the pathway of -1 frameshifting: Kinetics.Biochem Biophys Rep. 2016 Feb 10;5:453-467. doi: 10.1016/j.bbrep.2016.02.008. eCollection 2016 Mar. Biochem Biophys Rep. 2016. PMID: 28955853 Free PMC article.
-
The first gapless, reference-quality, fully annotated genome from a Southern Han Chinese individual.G3 (Bethesda). 2023 Mar 9;13(3):jkac321. doi: 10.1093/g3journal/jkac321. G3 (Bethesda). 2023. PMID: 36630290 Free PMC article.
References
-
- Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461(7268):1234–1242. - PubMed
-
- Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. - PubMed
-
- Sydow JF, Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr Opin Struct Biol. 2009;19(6):732–739. - PubMed
-
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136(4):763–776. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

