Modeling evolution of spatially distributed bacterial communities: a simulation with the haploid evolutionary constructor
- PMID: 25708911
- PMCID: PMC4331802
- DOI: 10.1186/1471-2148-15-S1-S3
Modeling evolution of spatially distributed bacterial communities: a simulation with the haploid evolutionary constructor
Abstract
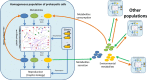
Background: Multiscale approaches for integrating submodels of various levels of biological organization into a single model became the major tool of systems biology. In this paper, we have constructed and simulated a set of multiscale models of spatially distributed microbial communities and study an influence of unevenly distributed environmental factors on the genetic diversity and evolution of the community members.
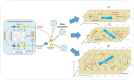
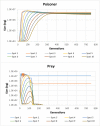
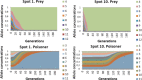
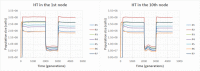
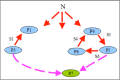
Results: Haploid Evolutionary Constructor software http://evol-constructor.bionet.nsc.ru/ was expanded by adding the tool for the spatial modeling of a microbial community (1D, 2D and 3D versions). A set of the models of spatially distributed communities was built to demonstrate that the spatial distribution of cells affects both intensity of selection and evolution rate.
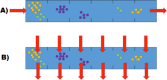
Conclusion: In spatially heterogeneous communities, the change in the direction of the environmental flow might be reflected in local irregular population dynamics, while the genetic structure of populations (frequencies of the alleles) remains stable. Furthermore, in spatially heterogeneous communities, the chemotaxis might dramatically affect the evolution of community members.
Figures





References
-
- Zavarzin G. In: Glob Ecol. 1. Jørgensen SE, editor. Academic Press; 2010. Microbial Cycles; pp. 183–190.
-
- Dobretsov N, Kolchanov N, Rozanov A, Zavarzin G. Biosphere Origin and Evolution. Boston, MA: Springer US; 2008. p. 426.
-
- Madigan MT, Martinko JM, Bender KS, Buckley DH, Stahl DA. Brock Biology of Microorganisms. 14. New York: Benjamin Cummings; 2014. p. 1136.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

