A small RNA mediated regulation of a stress-activated retrotransposon and the tissue specific transposition during the reproductive period in Arabidopsis
- PMID: 25709612
- PMCID: PMC4321352
- DOI: 10.3389/fpls.2015.00048
A small RNA mediated regulation of a stress-activated retrotransposon and the tissue specific transposition during the reproductive period in Arabidopsis
Abstract
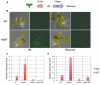
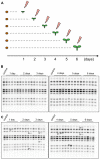
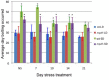
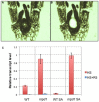
Transposable elements (TEs) are key elements that facilitate genome evolution of the host organism. A number of studies have assessed the functions of TEs, which change gene expression in the host genome. Activation of TEs is controlled by epigenetic modifications such as DNA methylation and histone modifications. Several recent studies have reported that TEs can also be activated by biotic or abiotic stress in some plants. We focused on a Ty1/copia retrotransposon, ONSEN, that is activated by heat stress (HS) in Arabidopsis. We found that transcriptional activation of ONSEN was regulated by a small interfering RNA (siRNA)-related pathway, and the activation could also be induced by oxidative stress. Mutants deficient in siRNA biogenesis that were exposed to HS at the initial stages of vegetative growth showed transgenerational transposition. The transposition was also detected in the progeny, which originated from tissue that had differentiated after exposure to the HS. The results indicated that in some undifferentiated cells, transpositional activity could be maintained quite long after exposure to the HS.
Keywords: Arabidopsis thaliana; ONSEN; environmental stress; small RNA; transposon.
Figures






Similar articles
-
The effects of heat induction and the siRNA biogenesis pathway on the transgenerational transposition of ONSEN, a copia-like retrotransposon in Arabidopsis thaliana.Plant Cell Physiol. 2012 May;53(5):824-33. doi: 10.1093/pcp/pcr179. Epub 2011 Dec 14. Plant Cell Physiol. 2012. PMID: 22173101
-
ONSEN shows different transposition activities in RdDM pathway mutants.Genes Genet Syst. 2020 Oct 23;95(4):183-190. doi: 10.1266/ggs.20-00019. Epub 2020 Sep 5. Genes Genet Syst. 2020. PMID: 32893196
-
Inducible Transposition of a Heat-Activated Retrotransposon in Tissue Culture.Plant Cell Physiol. 2017 Feb 1;58(2):375-384. doi: 10.1093/pcp/pcw202. Plant Cell Physiol. 2017. PMID: 28013279
-
Small RNAs and transposon silencing in plants.Dev Growth Differ. 2012 Jan;54(1):100-7. doi: 10.1111/j.1440-169X.2011.01309.x. Epub 2011 Dec 12. Dev Growth Differ. 2012. PMID: 22150226 Review.
-
[Molecular mechanisms of genetic transposition inhibition by piRNA].Yi Chuan. 2018 Jun 20;40(6):445-450. doi: 10.16288/j.yczz.18-072. Yi Chuan. 2018. PMID: 29959117 Review. Chinese.
Cited by
-
m6A RNA demethylase AtALKBH9B promotes mobilization of a heat-activated long terminal repeat retrotransposon in Arabidopsis.Sci Adv. 2023 Dec;9(48):eadf3292. doi: 10.1126/sciadv.adf3292. Epub 2023 Nov 29. Sci Adv. 2023. PMID: 38019921 Free PMC article.
-
Transposable elements underlie genetic adaptation.Nat Plants. 2024 Nov;10(11):1617-1618. doi: 10.1038/s41477-024-01792-y. Nat Plants. 2024. PMID: 39333350 No abstract available.
-
Comparative genomics of muskmelon reveals a potential role for retrotransposons in the modification of gene expression.Commun Biol. 2020 Aug 13;3(1):432. doi: 10.1038/s42003-020-01172-0. Commun Biol. 2020. PMID: 32792560 Free PMC article.
-
Experimentally heat-induced transposition increases drought tolerance in Arabidopsis thaliana.New Phytol. 2022 Oct;236(1):182-194. doi: 10.1111/nph.18322. Epub 2022 Jul 9. New Phytol. 2022. PMID: 35715973 Free PMC article.
-
A Stress-Activated Transposon in Arabidopsis Induces Transgenerational Abscisic Acid Insensitivity.Sci Rep. 2016 Mar 15;6:23181. doi: 10.1038/srep23181. Sci Rep. 2016. PMID: 26976262 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

