No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales
- PMID: 25710183
- PMCID: PMC4542034
- DOI: 10.1038/ismej.2015.20
No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales
Abstract
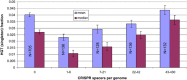
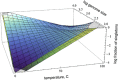
The CRISPR (clustered, regularly, interspaced, short, palindromic repeats)-Cas (CRISPR-associated genes) systems of archaea and bacteria provide adaptive immunity against viruses and other selfish elements and are believed to curtail horizontal gene transfer (HGT). Limiting acquisition of new genetic material could be one of the sources of the fitness cost of CRISPR-Cas maintenance and one of the causes of the patchy distribution of CRISPR-Cas among bacteria, and across environments. We sought to test the hypothesis that the activity of CRISPR-Cas in microbes is negatively correlated with the extent of recent HGT. Using three independent measures of HGT, we found no significant dependence between the length of CRISPR arrays, which reflects the activity of the immune system, and the estimated number of recent HGT events. In contrast, we observed a significant negative dependence between the estimated extent of HGT and growth temperature of microbes, which could be explained by the lower genetic diversity in hotter environments. We hypothesize that the relevant events in the evolution of resistance to mobile elements and proclivity for HGT, to which CRISPR-Cas systems seem to substantially contribute, occur on the population scale rather than on the timescale of species evolution.
Figures

References
-
- Anderson RE, Brazelton WJ, Baross JA. Using CRISPRs as a metagenomic tool to identify microbial hosts of a diffuse flow hydrothermal vent viral assemblage. FEMS Microbiol Ecol. 2011;77:120–133. - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
-
- Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12:177–186. - PubMed
-
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

