Structural basis of activity against aztreonam and extended spectrum cephalosporins for two carbapenem-hydrolyzing class D β-lactamases from Acinetobacter baumannii
- PMID: 25710192
- PMCID: PMC4476283
- DOI: 10.1021/bi501547k
Structural basis of activity against aztreonam and extended spectrum cephalosporins for two carbapenem-hydrolyzing class D β-lactamases from Acinetobacter baumannii
Abstract
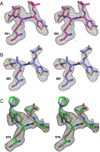
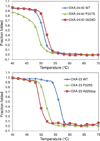
The carbapenem-hydrolyzing class D β-lactamases OXA-23 and OXA-24/40 have emerged worldwide as causative agents for β-lactam antibiotic resistance in Acinetobacter species. Many variants of these enzymes have appeared clinically, including OXA-160 and OXA-225, which both contain a P → S substitution at homologous positions in the OXA-24/40 and OXA-23 backgrounds, respectively. We purified OXA-160 and OXA-225 and used steady-state kinetic analysis to compare the substrate profiles of these variants to their parental enzymes, OXA-24/40 and OXA-23. OXA-160 and OXA-225 possess greatly enhanced hydrolytic activities against aztreonam, ceftazidime, cefotaxime, and ceftriaxone when compared to OXA-24/40 and OXA-23. These enhanced activities are the result of much lower Km values, suggesting that the P → S substitution enhances the binding affinity of these drugs. We have determined the structures of the acylated forms of OXA-160 (with ceftazidime and aztreonam) and OXA-225 (ceftazidime). These structures show that the R1 oxyimino side-chain of these drugs occupies a space near the β5-β6 loop and the omega loop of the enzymes. The P → S substitution found in OXA-160 and OXA-225 results in a deviation of the β5-β6 loop, relieving the steric clash with the R1 side-chain carboxypropyl group of aztreonam and ceftazidime. These results reveal worrying trends in the enhancement of substrate spectrum of class D β-lactamases but may also provide a map for β-lactam improvement.
Figures


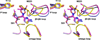
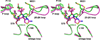



References
-
- Evans BA, Hamouda A, Amyes SG. The rise of carbapenem-resistant Acinetobacter baumannii. Curr. Pharm. Des. 2013;19:223–238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

