Substrate pathways in the nitrogenase MoFe protein by experimental identification of small molecule binding sites
- PMID: 25710326
- PMCID: PMC4590346
- DOI: 10.1021/bi501313k
Substrate pathways in the nitrogenase MoFe protein by experimental identification of small molecule binding sites
Abstract
In the nitrogenase molybdenum-iron (MoFe) protein, we have identified five potential substrate access pathways from the protein surface to the FeMo-cofactor (the active site) or the P-cluster using experimental structures of Xe pressurized into MoFe protein crystals from Azotobacter vinelandii and Clostridium pasteurianum. Additionally, all published structures of the MoFe protein, including those from Klebsiella pneumoniae, were analyzed for the presence of nonwater, small molecules bound to the protein interior. Each pathway is based on identification of plausible routes from buried small molecule binding sites to both the protein surface and a metallocluster. Of these five pathways, two have been previously suggested as substrate access pathways. While the small molecule binding sites are not conserved among the three species of MoFe protein, residues lining the pathways are generally conserved, indicating that the proposed pathways may be accessible in all three species. These observations imply that there is unlikely a unique pathway utilized for substrate access from the protein surface to the active site; however, there may be preferred pathways such as those described here.
Figures
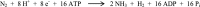

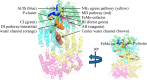



Similar articles
-
Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08 Å resolution: comparison with the Azotobacter vinelandii MoFe protein.Acta Crystallogr D Biol Crystallogr. 2015 Feb;71(Pt 2):274-82. doi: 10.1107/S1399004714025243. Epub 2015 Jan 23. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25664737 Free PMC article.
-
Formation of a tight 1:1 complex of Clostridium pasteurianum Fe protein-Azotobacter vinelandii MoFe protein: evidence for long-range interactions between the Fe protein binding sites during catalytic hydrogen evolution.Biochemistry. 2000 Sep 19;39(37):11434-40. doi: 10.1021/bi0002939. Biochemistry. 2000. PMID: 10985789
-
X-ray crystal structure of the nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 3.0-A resolution.Biochemistry. 1993 Jul 20;32(28):7104-15. doi: 10.1021/bi00079a006. Biochemistry. 1993. PMID: 8393705
-
Identification of a nitrogenase protein-protein interaction site defined by residues 59 through 67 within the Azotobacter vinelandii Fe protein.J Biol Chem. 1994 Nov 11;269(45):28076-83. J Biol Chem. 1994. PMID: 7961744
-
A substrate channel in the nitrogenase MoFe protein.J Biol Inorg Chem. 2009 Sep;14(7):1015-22. doi: 10.1007/s00775-009-0544-2. Epub 2009 May 21. J Biol Inorg Chem. 2009. PMID: 19458968 Free PMC article.
Cited by
-
Small-Molecule Tunnels in Metalloenzymes Viewed as Extensions of the Active Site.Acc Chem Res. 2021 May 4;54(9):2185-2195. doi: 10.1021/acs.accounts.1c00058. Epub 2021 Apr 22. Acc Chem Res. 2021. PMID: 33886257 Free PMC article. Review.
-
Structural evolution of nitrogenase states under alkaline turnover.Nat Commun. 2024 Dec 2;15(1):10472. doi: 10.1038/s41467-024-54713-0. Nat Commun. 2024. PMID: 39622820 Free PMC article.
-
Nitrogenase beyond the Resting State: A Structural Perspective.Molecules. 2023 Dec 5;28(24):7952. doi: 10.3390/molecules28247952. Molecules. 2023. PMID: 38138444 Free PMC article. Review.
-
Mutational Analysis of the Nitrogenase Carbon Monoxide Protective Protein CowN Reveals That a Conserved C-Terminal Glutamic Acid Residue Is Necessary for Its Activity.Biochemistry. 2024 Jan 2;63(1):152-158. doi: 10.1021/acs.biochem.3c00421. Epub 2023 Dec 13. Biochemistry. 2024. PMID: 38091601 Free PMC article.
-
CryoEM-sampling of metastable conformations appearing in cofactor-ligand association and catalysis of glutamate dehydrogenase.Sci Rep. 2024 May 15;14(1):11165. doi: 10.1038/s41598-024-61793-x. Sci Rep. 2024. PMID: 38750092 Free PMC article.
References
-
- Erisman J. W.; Sutton M. A.; Galloway J.; Klimont Z.; Winiwarter W. (2008) How a century of ammonia synthesis changed the world. Nature Geosci. 1, 636–639.
-
- Kitano M.; Inoue Y.; Yamazaki Y.; Hayashi F.; Kanbara S.; Matsuishi S.; Yokoyama T.; Kim S.-W.; Hara M.; Hosono H. (2012) Ammonia synthesis using a stable electrode as an electron donor and reversible hydrogen store. Nat. Chem. 4, 934–940. - PubMed
-
- Hartwig J. F. (2010) Organotransition Metal Chemistry: From Bonding to Catalysis, p 540, University Science Books, Mill Valley, CA.
-
- Rees D. C.; Howard J. B. (2000) Nitrogenase: Standing at the crossroads. Curr. Opin. Chem. Biol. 4, 559–566. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

