Visualizing transient dark states by NMR spectroscopy
- PMID: 25710841
- PMCID: PMC6276111
- DOI: 10.1017/S0033583514000122
Visualizing transient dark states by NMR spectroscopy
Abstract
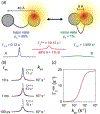
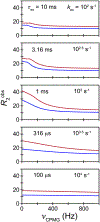
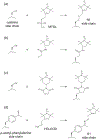
Myriad biological processes proceed through states that defy characterization by conventional atomic-resolution structural biological methods. The invisibility of these 'dark' states can arise from their transient nature, low equilibrium population, large molecular weight, and/or heterogeneity. Although they are invisible, these dark states underlie a range of processes, acting as encounter complexes between proteins and as intermediates in protein folding and aggregation. New methods have made these states accessible to high-resolution analysis by nuclear magnetic resonance (NMR) spectroscopy, as long as the dark state is in dynamic equilibrium with an NMR-visible species. These methods - paramagnetic NMR, relaxation dispersion, saturation transfer, lifetime line broadening, and hydrogen exchange - allow the exploration of otherwise invisible states in exchange with a visible species over a range of timescales, each taking advantage of some unique property of the dark state to amplify its effect on a particular NMR observable. In this review, we introduce these methods and explore two specific techniques - paramagnetic relaxation enhancement and dark state exchange saturation transfer - in greater detail.
Figures








References
-
- ALTENBACH C, OH KJ, TRABANINO RJ, HIDEG K & HUBBELL WL (2001). Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: experimental strategies and practical limitations. Biochemistry 40, 15471–15482. - PubMed
-
- BABU YS, SACK JS, GREENHOUGH TJ, BUGG CE, MEANS AR & COOK WJ (1985). Three-dimensional structure of calmodulin. Nature 315, 37–40. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

