Traffic lines: new tools for genetic analysis in Arabidopsis thaliana
- PMID: 25711279
- PMCID: PMC4423376
- DOI: 10.1534/genetics.114.173435
Traffic lines: new tools for genetic analysis in Arabidopsis thaliana
Abstract
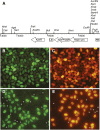
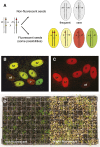
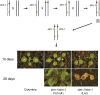
Genetic analysis requires the ability to identify the genotypes of individuals in a segregating population. This task is straightforward if each genotype has a distinctive phenotype, but is difficult if these genotypes are phenotypically similar or identical. We show that Arabidopsis seeds homozygous or heterozygous for a mutation of interest can be identified in a segregating family by placing the mutation in trans to a chromosome carrying a pair of seed-expressed green and red fluorescent transgenes (a "traffic line") that flank the mutation. Nonfluorescent seeds in the self-pollinated progeny of such a heterozygous plant are usually homozygous for the mutation, whereas seeds with intermediate green and red fluorescence are typically heterozygous for the mutation. This makes it possible to identify seedlings homozygous for mutations that lack an obvious seedling phenotype, and also facilitates the analysis of lethal or sterile mutations, which must be propagated in heterozygous condition. Traffic lines can also be used to identify progeny that have undergone recombination within a defined region of the genome, facilitating genetic mapping and the production of near-isogenic lines. We produced 488 transgenic lines containing single genome-mapped insertions of NAP:dsRED and NAP:eGFP in Columbia (330 lines) and Landsberg erecta (158 lines) and generated sets of traffic lines that span most regions of the Arabidopsis genome. We demonstrated the utility of these lines for identifying seeds of a specific genotype and for generating near-isogenic lines using mutations of WUSCHEL and SHOOTMERISTEMLESS. This new resource significantly decreases the effort and cost of genotyping segregating families and increases the efficiency of experiments that rely on the ability to detect recombination in a defined chromosomal segment.
Keywords: T-DNA; mapping; recombination; stm; wus.
Copyright © 2015 by the Genetics Society of America.
Figures



Similar articles
-
Short-interval traffic lines: versatile tools for genetic analysis in Arabidopsis thaliana.G3 (Bethesda). 2022 Sep 30;12(10):jkac202. doi: 10.1093/g3journal/jkac202. G3 (Bethesda). 2022. PMID: 36018241 Free PMC article.
-
A new seed-based assay for meiotic recombination in Arabidopsis thaliana.Plant J. 2005 Aug;43(3):458-66. doi: 10.1111/j.1365-313X.2005.02466.x. Plant J. 2005. PMID: 16045480
-
Reverse breeding in Arabidopsis thaliana generates homozygous parental lines from a heterozygous plant.Nat Genet. 2012 Mar 11;44(4):467-70. doi: 10.1038/ng.2203. Nat Genet. 2012. PMID: 22406643
-
Mapping of BnMs4 and BnRf to a common microsyntenic region of Arabidopsis thaliana chromosome 3 using intron polymorphism markers.Theor Appl Genet. 2012 May;124(7):1193-200. doi: 10.1007/s00122-011-1779-1. Epub 2012 Jan 14. Theor Appl Genet. 2012. PMID: 22246313 Review.
-
Molecular neuroanatomy's "Three Gs": a primer.Neuron. 2007 Apr 5;54(1):17-34. doi: 10.1016/j.neuron.2007.03.009. Neuron. 2007. PMID: 17408575 Free PMC article. Review.
Cited by
-
An siRNA-guided ARGONAUTE protein directs RNA polymerase V to initiate DNA methylation.Nat Plants. 2021 Nov;7(11):1461-1474. doi: 10.1038/s41477-021-01008-7. Epub 2021 Nov 8. Nat Plants. 2021. PMID: 34750500 Free PMC article.
-
Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana.PLoS Genet. 2018 Apr 19;14(4):e1007337. doi: 10.1371/journal.pgen.1007337. eCollection 2018 Apr. PLoS Genet. 2018. PMID: 29672610 Free PMC article.
-
KinG Is a Plant-Specific Kinesin That Regulates Both Intra- and Intercellular Movement of SHORT-ROOT.Plant Physiol. 2018 Jan;176(1):392-405. doi: 10.1104/pp.17.01518. Epub 2017 Nov 9. Plant Physiol. 2018. PMID: 29122988 Free PMC article.
-
Arabidopsis HEAT SHOCK FACTOR BINDING PROTEIN is required to limit meiotic crossovers and HEI10 transcription.EMBO J. 2022 Jul 18;41(14):e109958. doi: 10.15252/embj.2021109958. Epub 2022 Jun 7. EMBO J. 2022. PMID: 35670129 Free PMC article.
-
The effect of DNA polymorphisms and natural variation on crossover hotspot activity in Arabidopsis hybrids.Nat Commun. 2023 Jan 3;14(1):33. doi: 10.1038/s41467-022-35722-3. Nat Commun. 2023. PMID: 36596804 Free PMC article.
References
-
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., et al. , 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. - PubMed
-
- Ashburner M., Golic K., Hawley R. S., 2005. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Barton M. K., Poethig R. S., 1993. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shootmeristemless mutant. Development 119: 823–831.
-
- Berchowitz L. E., Copenhaver G. P., 2008. Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nat. Protoc. 3: 41–50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

