Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages
- PMID: 25712094
- PMCID: PMC4357732
- DOI: 10.1093/nar/gkv143
Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages
Abstract
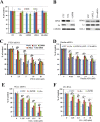
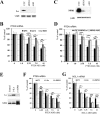
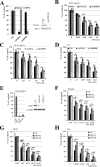
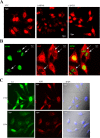
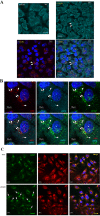
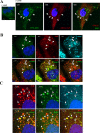
Although the RNase H-dependent mechanism of inhibition of gene expression by chemically modified antisense oligonucleotides (ASOs) has been well characterized, little is known about the interactions between ASOs and intracellular proteins that may alter cellular localization and/or potency of ASOs. Here, we report the identification of 56 intracellular ASO-binding proteins using multi-step affinity selection approaches. Many of the tested proteins had no significant effect on ASO activity; however, some proteins, including La/SSB, NPM1, ANXA2, VARS and PC4, appeared to enhance ASO activities, likely through mechanisms related to subcellular distribution. VARS and ANXA2 co-localized with ASOs in endocytic organelles, and reduction in the level of VARS altered lysosome/ASO localization patterns, implying that these proteins may facilitate ASO release from the endocytic pathway. Depletion of La and NPM1 reduced nuclear ASO levels, suggesting potential roles in ASO nuclear accumulation. On the other hand, Ku70 and Ku80 proteins inhibited ASO activity, most likely by competition with RNase H1 for ASO/RNA duplex binding. Our results demonstrate that phosphorothioate-modified ASOs bind a set of cellular proteins that affect ASO activity via different mechanisms.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Lima W., Wu H., Crooke S.T. The RNase H Mechanism. In: Crooke ST, editor. Antisense Drug Technology—Principles, Strategies, and Applications. 2nd edn. Boca Raton, FL: CRC Press; 2008. pp. 47–74.
-
- Crooke S.T., Vickers T.A., Lima W.F., Wu H.-J. Mechanisms of Antisense Drug Action, an Introduction. In: Crooke ST, editor. Antisense Drug Technology—Principles, Strategies, and Applications. 2nd edn. Boca Raton, FL: CRC Press; 2008. pp. 3–46.
-
- Nowotny M., Gaidamakov S.A., Crouch R.J., Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. - PubMed
-
- Wu H., Lima W.F., Zhang H., Fan A., Sun H., Crooke S.T. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

