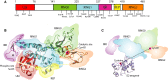
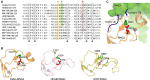
Parkin structure and function
- PMID: 25712550
- PMCID: PMC4672691
- DOI: 10.1111/febs.13249
Parkin structure and function
Abstract
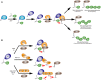
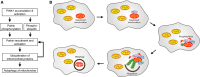
Mutations in the parkin or PINK1 genes are the leading cause of the autosomal recessive form of Parkinson's disease. The gene products, the E3 ubiquitin ligase parkin and the serine/threonine kinase PINK1, are neuroprotective proteins, which act together in a mitochondrial quality control pathway. Here, we review the structure of parkin and mechanisms of its autoinhibition and function as a ubiquitin ligase. We present a model for the recruitment and activation of parkin as a key regulatory step in the clearance of depolarized or damaged mitochondria by autophagy (mitophagy). We conclude with a brief overview of other functions of parkin and considerations for drug discovery in the mitochondrial quality control pathway.
Keywords: PINK1; mitophagy; parkin; ubiquitin; ubiquitination.
© 2015 The Authors. FEBS Journal published by John Wiley & Sons Ltd on behalf of FEBS.
Figures
References
-
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

