Key residues in the nicotinic acetylcholine receptor β2 subunit contribute to α-conotoxin LvIA binding
- PMID: 25713061
- PMCID: PMC4392282
- DOI: 10.1074/jbc.M114.632646
Key residues in the nicotinic acetylcholine receptor β2 subunit contribute to α-conotoxin LvIA binding
Abstract
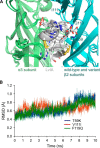
α-Conotoxin LvIA (α-CTx LvIA) is a small peptide from the venom of the carnivorous marine gastropod Conus lividus and is the most selective inhibitor of α3β2 nicotinic acetylcholine receptors (nAChRs) known to date. It can distinguish the α3β2 nAChR subtype from the α6β2* (* indicates the other subunit) and α3β4 nAChR subtypes. In this study, we performed mutational studies to assess the influence of residues of the β2 subunit versus those of the β4 subunit on the binding of α-CTx LvIA. Although two β2 mutations, α3β2[F119Q] and α3β2[T59K], strongly enhanced the affinity of LvIA, the β2 mutation α3β2[V111I] substantially reduced the binding of LvIA. Increased activity of LvIA was also observed when the β2-T59L mutant was combined with the α3 subunit. There were no significant difference in inhibition of α3β2[T59I], α3β2[Q34A], and α3β2[K79A] nAChRs when compared with wild-type α3β2 nAChR. α-CTx LvIA displayed slower off-rate kinetics at α3β2[F119Q] and α3β2[T59K] than at the wild-type receptor, with the latter mutant having the most pronounced effect. Taken together, these data provide evidence that the β2 subunit contributes to α-CTx LvIA binding and selectivity. The results demonstrate that Val(111) is critical and facilitates LvIA binding; this position has not previously been identified as important to binding of other 4/7 framework α-conotoxins. Thr(59) and Phe(119) of the β2 subunit appear to interfere with LvIA binding, and their replacement by the corresponding residues of the β4 subunit leads to increased affinity.
Keywords: Docking; Mutant α3β2 Subtype.; Receptor; Receptor Structure-Function; Receptor-interacting Protein (RIP); Toxin; α-Conotoxin LvIA; α3β2 nAChR; β subunit contribution.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




Similar articles
-
Key Structural Determinants in the Agonist Binding Loops of Human β2 and β4 Nicotinic Acetylcholine Receptor Subunits Contribute to α3β4 Subtype Selectivity of α-Conotoxins.J Biol Chem. 2016 Nov 4;291(45):23779-23792. doi: 10.1074/jbc.M116.730804. Epub 2016 Sep 19. J Biol Chem. 2016. PMID: 27646000 Free PMC article.
-
A novel α4/7-conotoxin LvIA from Conus lividus that selectively blocks α3β2 vs. α6/α3β2β3 nicotinic acetylcholine receptors.FASEB J. 2014 Apr;28(4):1842-53. doi: 10.1096/fj.13-244103. Epub 2014 Jan 7. FASEB J. 2014. PMID: 24398291 Free PMC article.
-
Atypical alpha-conotoxin LtIA from Conus litteratus targets a novel microsite of the alpha3beta2 nicotinic receptor.J Biol Chem. 2010 Apr 16;285(16):12355-66. doi: 10.1074/jbc.M109.079012. Epub 2010 Feb 9. J Biol Chem. 2010. PMID: 20145249 Free PMC article.
-
α-Conotoxins active at α3-containing nicotinic acetylcholine receptors and their molecular determinants for selective inhibition.Br J Pharmacol. 2018 Jun;175(11):1855-1868. doi: 10.1111/bph.13852. Epub 2017 Jun 11. Br J Pharmacol. 2018. PMID: 28477355 Free PMC article. Review.
-
Mutagenesis of α-Conotoxins for Enhancing Activity and Selectivity for Nicotinic Acetylcholine Receptors.Toxins (Basel). 2019 Feb 13;11(2):113. doi: 10.3390/toxins11020113. Toxins (Basel). 2019. PMID: 30781866 Free PMC article. Review.
Cited by
-
Neuronal Nicotinic Acetylcholine Receptor Modulators from Cone Snails.Mar Drugs. 2018 Jun 13;16(6):208. doi: 10.3390/md16060208. Mar Drugs. 2018. PMID: 29899286 Free PMC article. Review.
-
Key Structural Determinants in the Agonist Binding Loops of Human β2 and β4 Nicotinic Acetylcholine Receptor Subunits Contribute to α3β4 Subtype Selectivity of α-Conotoxins.J Biol Chem. 2016 Nov 4;291(45):23779-23792. doi: 10.1074/jbc.M116.730804. Epub 2016 Sep 19. J Biol Chem. 2016. PMID: 27646000 Free PMC article.
-
Characterization of an α 4/7-Conotoxin LvIF from Conus lividus That Selectively Blocks α3β2 Nicotinic Acetylcholine Receptor.Mar Drugs. 2021 Jul 17;19(7):398. doi: 10.3390/md19070398. Mar Drugs. 2021. PMID: 34356823 Free PMC article.
-
Residues Responsible for the Selectivity of α-Conotoxins for Ac-AChBP or nAChRs.Mar Drugs. 2016 Oct 11;14(10):173. doi: 10.3390/md14100173. Mar Drugs. 2016. PMID: 27727162 Free PMC article. Review.
-
Interactions of the α3β2 Nicotinic Acetylcholine Receptor Interfaces with α-Conotoxin LsIA and its Carboxylated C-terminus Analogue: Molecular Dynamics Simulations.Mar Drugs. 2020 Jul 3;18(7):349. doi: 10.3390/md18070349. Mar Drugs. 2020. PMID: 32635340 Free PMC article.
References
-
- Wallace T. L., Bertrand D. (2013) Importance of the nicotinic acetylcholine receptor system in the prefrontal cortex. Biochem. Pharmacol. 85, 1713–1720 - PubMed
-
- Corringer P. J., Le Novère N., Changeux J. P. (2000) Nicotinic receptors at the amino acid level. Annu. Rev. Pharmacol. Toxicol. 40, 431–458 - PubMed
-
- Rahman S. (2013) Nicotinic receptors as therapeutic targets for drug addictive disorders. CNS Neurol. Disord. Drug Targets 12, 633–640 - PubMed
-
- Somm E. (2014) Nicotinic cholinergic signaling in adipose tissue and pancreatic islets biology: revisited function and therapeutic perspectives. Arch. Immunol. Ther. Exp. (Warsz.) 62, 87–101 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

