Chaperone-mediated 26S proteasome remodeling facilitates free K63 ubiquitin chain production and aggresome clearance
- PMID: 25713068
- PMCID: PMC4392251
- DOI: 10.1074/jbc.M114.627950
Chaperone-mediated 26S proteasome remodeling facilitates free K63 ubiquitin chain production and aggresome clearance
Abstract
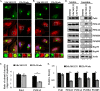
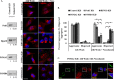
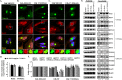
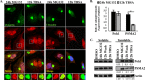
Efficient elimination of misfolded proteins by the proteasome system is critical for proteostasis. Inadequate proteasome capacity can lead to aberrant aggregation of misfolded proteins and inclusion body formation, a hallmark of neurodegenerative disease. The proteasome system cannot degrade aggregated proteins; however, it stimulates autophagy-dependent aggregate clearance by producing unanchored lysine (K)63-linked ubiquitin chains via the proteasomal deubiquitinating enzyme Poh1. The canonical function of Poh1, which removes ubiquitin chains en bloc from proteasomal substrates prior to their degradation, requires intact 26S proteasomes. Here we present evidence that during aggresome clearance, 20S proteasomes dissociate from protein aggregates, while Poh1 and selective subunits of 19S proteasomes are retained. The dissociation of 20S proteasome components requires the molecular chaperone Hsp90. Hsp90 inhibition suppresses 26S proteasome remodeling, unanchored ubiquitin chain production, and aggresome clearance. Our results suggest that 26S proteasomes undergo active remodeling to generate a Poh1-dependent K63-deubiquitinating enzyme to facilitate protein aggregate clearance.
Keywords: aggresome; autophagy; deubiquitylation (deubiquitination); histone deacetylase 6 (HDAC6); proteasome; ubiquitin.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Proteasomes activate aggresome disassembly and clearance by producing unanchored ubiquitin chains.Mol Cell. 2013 Sep 26;51(6):819-28. doi: 10.1016/j.molcel.2013.08.016. Epub 2013 Sep 12. Mol Cell. 2013. PMID: 24035499 Free PMC article.
-
UCH-L1 Inhibition Suppresses tau Aggresome Formation during Proteasomal Impairment.Mol Neurobiol. 2018 May;55(5):3812-3821. doi: 10.1007/s12035-017-0558-7. Epub 2017 May 24. Mol Neurobiol. 2018. PMID: 28540657
-
Deubiquitination by proteasome is coordinated with substrate translocation for proteolysis in vivo.Exp Cell Res. 2005 Jul 15;307(2):436-51. doi: 10.1016/j.yexcr.2005.03.031. Exp Cell Res. 2005. PMID: 15950624
-
Role of the aggresome pathway in cancer: targeting histone deacetylase 6-dependent protein degradation.Cancer Res. 2008 Apr 15;68(8):2557-60. doi: 10.1158/0008-5472.CAN-07-5989. Cancer Res. 2008. PMID: 18413721 Review.
-
[Histone deacetylase 6: the key regulator of misfolded proteins].Sheng Li Ke Xue Jin Zhan. 2010 Apr;41(2):112-6. Sheng Li Ke Xue Jin Zhan. 2010. PMID: 21416996 Review. Chinese.
Cited by
-
Interplay Between the Autophagy-Lysosomal Pathway and the Ubiquitin-Proteasome System: A Target for Therapeutic Development in Alzheimer's Disease.Front Cell Neurosci. 2018 May 9;12:126. doi: 10.3389/fncel.2018.00126. eCollection 2018. Front Cell Neurosci. 2018. PMID: 29867359 Free PMC article. Review.
-
K29-linked free polyubiquitin chains affect ribosome biogenesis and direct ribosomal proteins to the intranuclear quality control compartment.Mol Cell. 2024 Jun 20;84(12):2337-2352.e9. doi: 10.1016/j.molcel.2024.05.018. Epub 2024 Jun 12. Mol Cell. 2024. PMID: 38870935 Free PMC article.
-
Oligomerization-primed coiled-coil domain interaction with Ubc13 confers processivity to TRAF6 ubiquitin ligase activity.Nat Commun. 2017 Oct 9;8(1):814. doi: 10.1038/s41467-017-01290-0. Nat Commun. 2017. PMID: 28993672 Free PMC article.
-
A Single α Helix Drives Extensive Remodeling of the Proteasome Lid and Completion of Regulatory Particle Assembly.Cell. 2015 Oct 8;163(2):432-44. doi: 10.1016/j.cell.2015.09.022. Cell. 2015. PMID: 26451487 Free PMC article.
-
A Novel Dual HDAC6 and Tubulin Inhibitor, MPT0B451, Displays Anti-tumor Ability in Human Cancer Cells in Vitro and in Vivo.Front Pharmacol. 2018 Mar 13;9:205. doi: 10.3389/fphar.2018.00205. eCollection 2018. Front Pharmacol. 2018. PMID: 29593536 Free PMC article.
References
-
- Kopito R. R. (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530 - PubMed
-
- Soto C. (2003) Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 4, 49–60 - PubMed
-
- Kawaguchi Y., Kovacs J. J., McLaurin A., Vance J. M., Ito A., Yao T. P. (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727–738 - PubMed
-
- McNaught K. S., Shashidharan P., Perl D. P., Jenner P., Olanow C. W. (2002) Aggresome-related biogenesis of Lewy bodies. Eur. J. Neurosci. 16, 2136–2148 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

