Structure of a BAG6 (Bcl-2-associated athanogene 6)-Ubl4a (ubiquitin-like protein 4a) complex reveals a novel binding interface that functions in tail-anchored protein biogenesis
- PMID: 25713138
- PMCID: PMC4392246
- DOI: 10.1074/jbc.M114.631804
Structure of a BAG6 (Bcl-2-associated athanogene 6)-Ubl4a (ubiquitin-like protein 4a) complex reveals a novel binding interface that functions in tail-anchored protein biogenesis
Abstract
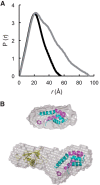
BAG6 is an essential protein that functions in two distinct biological pathways, ubiquitin-mediated protein degradation of defective polypeptides and tail-anchored (TA) transmembrane protein biogenesis in mammals, although its structural and functional properties remain unknown. We solved a crystal structure of the C-terminal heterodimerization domains of BAG6 and Ubl4a and characterized their interaction biochemically. Unexpectedly, the specificity and structure of the C terminus of BAG6, which was previously classified as a BAG domain, were completely distinct from those of the canonical BAG domain. Furthermore, the tight association of BAG6 and Ubl4a resulted in modulation of Ubl4a protein stability in cells. Therefore, we propose to designate the Ubl4a-binding region of BAG6 as the novel BAG-similar (BAGS) domain. The structure of Ubl4a, which interacts with BAG6, is similar to the yeast homologue Get5, which forms a homodimer. These observations indicate that the BAGS domain of BAG6 promotes the TA protein biogenesis pathway in mammals by the interaction with Ubl4a.
Keywords: Membrane Protein; Protein Assembly; Small-angle X-ray Scattering (SAXS); Tail Anchor; Ubiquitin; X-ray Crystallography.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Lüders J., Demand J., Höhfeld J. (2000) The ubiquitin-related BAG-1 provides a link between the molecular chaperone Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 275, 4613–4617 - PubMed
-
- Demand J., Alberti S., Patterson C., Höhfeld J. (2001) Cooperation of a ubiquitin domain protein and E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 11, 1569–1577 - PubMed
-
- Takayama S., Reed J. C. (2001) Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell Biol. 3, E237–E241 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

