The role of BPTF in melanoma progression and in response to BRAF-targeted therapy
- PMID: 25713167
- PMCID: PMC4555639
- DOI: 10.1093/jnci/djv034
The role of BPTF in melanoma progression and in response to BRAF-targeted therapy
Abstract
Background: Bromodomain PHD finger transcription factor (BPTF) plays an important role in chromatin remodeling, but its functional role in tumor progression is incompletely understood. Here we explore the oncogenic effects of BPTF in melanoma.
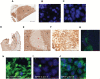
Methods: The consequences of differential expression of BPTF were explored using shRNA-mediated knockdown in several melanoma cell lines. Immunoblotting was used to assess the expression of various proteins regulated by BPTF. The functional role of BPTF in melanoma progression was investigated using assays of colony formation, invasion, cell cycle, sensitivity to selective BRAF inhibitors, and in xenograft models of melanoma progression (n = 12 mice per group). The biomarker role of BPTF in melanoma progression was assessed using fluorescence in situ hybridization and immunohistochemical analyses. All statistical tests were two-sided.
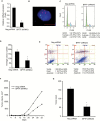
Results: shRNA-mediated BPTF silencing suppressed the proliferative capacity (by 65.5%) and metastatic potential (by 66.4%) of melanoma cells. Elevated BPTF copy number (mean ≥ 3) was observed in 28 of 77 (36.4%) melanomas. BPTF overexpression predicted poor survival in a cohort of 311 melanoma patients (distant metastasis-free survival P = .03, and disease-specific survival P = .008), and promoted resistance to BRAF inhibitors in melanoma cell lines. Metastatic melanoma tumors progressing on BRAF inhibitors contained low BPTF-expressing, apoptotic tumor cell subclones, indicating the continued presence of drug-responsive subclones within tumors demonstrating overall resistance to anti-BRAF agents.
Conclusions: These studies demonstrate multiple protumorigenic functions for BPTF and identify it as a novel target for anticancer therapy. They also suggest the combination of BPTF targeting with BRAF inhibitors as a novel therapeutic strategy for melanomas with mutant BRAF.
© The Author 2015. Published by Oxford University Press.
Figures




References
-
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78;273–304. - PubMed
-
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83(6):1011–1020. - PubMed
-
- Xiao H, Sandaltzopoulos R, Wang HM, et al. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8(3):531–543. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

