Thiopeptide antibiotics stimulate biofilm formation in Bacillus subtilis
- PMID: 25713360
- PMCID: PMC4364203
- DOI: 10.1073/pnas.1414272112
Thiopeptide antibiotics stimulate biofilm formation in Bacillus subtilis
Abstract
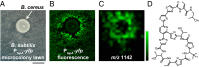
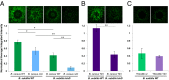
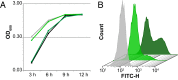
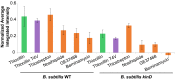
Bacteria have evolved the ability to produce a wide range of structurally complex natural products historically called "secondary" metabolites. Although some of these compounds have been identified as bacterial communication cues, more frequently natural products are scrutinized for antibiotic activities that are relevant to human health. However, there has been little regard for how these compounds might otherwise impact the physiology of neighboring microbes present in complex communities. Bacillus cereus secretes molecules that activate expression of biofilm genes in Bacillus subtilis. Here, we use imaging mass spectrometry to identify the thiocillins, a group of thiazolyl peptide antibiotics, as biofilm matrix-inducing compounds produced by B. cereus. We found that thiocillin increased the population of matrix-producing B. subtilis cells and that this activity could be abolished by multiple structural alterations. Importantly, a mutation that eliminated thiocillin's antibiotic activity did not affect its ability to induce biofilm gene expression in B. subtilis. We go on to show that biofilm induction appears to be a general phenomenon of multiple structurally diverse thiazolyl peptides and use this activity to confirm the presence of thiazolyl peptide gene clusters in other bacterial species. Our results indicate that the roles of secondary metabolites initially identified as antibiotics may have more complex effects--acting not only as killing agents, but also as specific modulators of microbial cellular phenotypes.
Keywords: Bacillus cereus; Bacillus subtilis; biofilm formation; thiazolyl antibiotics; thiopeptides.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Straight PD, Kolter R. Interspecies chemical communication in bacterial development. Annu Rev Microbiol. 2009;63:99–118. - PubMed
-
- Ryan RP, Dow JM. Diffusible signals and interspecies communication in bacteria. Microbiology. 2008;154(Pt 7):1845–1858. - PubMed
-
- Fajardo A, Martínez JL. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol. 2008;11(2):161–167. - PubMed
-
- Williams P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology. 2007;153(Pt 12):3923–3938. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

