Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity
- PMID: 25714049
- PMCID: PMC4376664
- DOI: 10.1038/nn.3975
Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity
Abstract
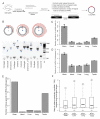
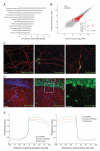
Circular RNAs (circRNAs) have re-emerged as an interesting RNA species. Using deep RNA profiling in different mouse tissues, we observed that circRNAs were substantially enriched in brain and a disproportionate fraction of them were derived from host genes that encode synaptic proteins. Moreover, on the basis of separate profiling of the RNAs localized in neuronal cell bodies and neuropil, circRNAs were, on average, more enriched in the neuropil than their host gene mRNA isoforms. Using high-resolution in situ hybridization, we visualized circRNA punctae in the dendrites of neurons. Consistent with the idea that circRNAs might regulate synaptic function during development, many circRNAs changed their abundance abruptly at a time corresponding to synaptogenesis. In addition, following a homeostatic downscaling of neuronal activity many circRNAs exhibited substantial up- or downregulation. Together, our data indicate that brain circRNAs are positioned to respond to and regulate synaptic function.
Figures


Comment in
-
Gear Up in Circles.Mol Cell. 2015 Jun 4;58(5):715-7. doi: 10.1016/j.molcel.2015.05.027. Mol Cell. 2015. PMID: 26046645
References
-
- Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi:nature11993 [pii] 10.1038/nature11993. - PubMed
-
- Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi:nature11928 [pii] 10.1038/nature11928. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

