Ultrastructural relationship of the phagophore with surrounding organelles
- PMID: 25714487
- PMCID: PMC4502653
- DOI: 10.1080/15548627.2015.1017178
Ultrastructural relationship of the phagophore with surrounding organelles
Abstract
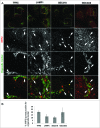
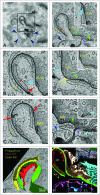
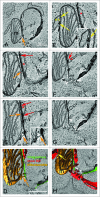
Phagophore nucleates from a subdomain of the endoplasmic reticulum (ER) termed the omegasome and also makes contact with other organelles such as mitochondria, Golgi complex, plasma membrane and recycling endosomes during its formation. We have used serial block face scanning electron microscopy (SB-EM) and electron tomography (ET) to image phagophore biogenesis in 3 dimensions and to determine the relationship between the phagophore and surrounding organelles at high resolution. ET was performed to confirm whether membrane contact sites (MCSs) are evident between the phagophore and those surrounding organelles. In addition to the known contacts with the ER, we identified MCSs between the phagophore and membranes from putative ER exit sites, late endosomes or lysosomes, the Golgi complex and mitochondria. We also show that one phagophore can have simultaneous MCSs with more than one organelle. Future membrane flux experiments are needed to determine whether membrane contacts also signify lipid translocation.
Keywords: 3D, 3 dimensional; ATG, autophagy-related; BSA, bovine serum albumin; COPII, coat protein II; ER, endoplasmic reticulum; ET, electron tomography; GOLGA2/GM130, golgin A2; Golgi complex; LAMP1, lysosomal-associated membrane protein 1; MAP1LC3/LC3, microtubule-associated protein 1 light chain 3; MCS, membrane contact site; PBS, phosphate-buffered saline; SB-EM, serial block-face scanning electron microscopy; SEC31A, SEC31 homolog A (S. cerevisiae); TFRC, transferrin receptor; WIPI2, WD repeat domain, phosphoinositide interacting 2; autophagy; electron tomography; immunoEM; immunoEM, immuno electron microscopy; lysosome; mitochondrion; serial block face scanning electron microscopy; three dimensional.
Figures


References
-
- Chan DC. Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol 2006; 22:79-99; PMID:16704336 - PubMed
-
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 2001; 20:5971-81; PMID:11689437; http://dx.doi.org/10.1093/emboj/20.21.5971 - DOI - PMC - PubMed
-
- Suzuki K, Ohsumi Y. Current knowledge of the pre-autophagosomal structure (PAS). FEBS Lett 2010; 584:1280-6; PMID:20138172; http://dx.doi.org/10.1016/j.febslet.2010.02.001 - DOI - PubMed
-
- Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci 2005; 118:7-18; PMID:15615779; http://dx.doi.org/10.1242/jcs.01620 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
