Highly stable and sensitive fluorescent probes (LysoProbes) for lysosomal labeling and tracking
- PMID: 25715948
- PMCID: PMC4341211
- DOI: 10.1038/srep08576
Highly stable and sensitive fluorescent probes (LysoProbes) for lysosomal labeling and tracking
Abstract
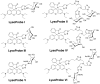
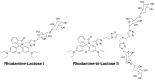
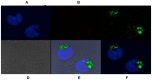
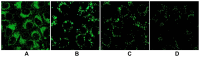
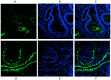
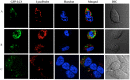
We report the design, synthesis and application of several new fluorescent probes (LysoProbes I-VI) that facilitate lysosomal pH monitoring and characterization of lysosome-dependent apoptosis. LysoProbes are superior to commercially available lysosome markers since the fluorescent signals are both stable and highly selective, and they will aid in characterization of lysosome morphology and trafficking. We predict that labeling of cancer cells and solid tumor tissues with LysoProbes will provide an important new tool for monitoring the role of lysosome trafficking in cancer invasion and metastasis.
Figures



References
-
- Maes H. & Agostinis P. Autophagy and mitophagy interplay in melanoma progression. Mitochondrion 17, S1567–7249 (2014). - PubMed
-
- Hönscheid P., Datta K. & Muders M. H. Autophagy: detection, regulation and its role in cancer and therapy response. Int J Radiat Biol. 90, 628–35 (2014). - PubMed
-
- He X., Li J., An S. & Jiang C. pH-sensitive drug-delivery systems for tumor targeting. Ther Deliv. 4, 1499–510 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

