Three-dimensional topology of the SMC2/SMC4 subcomplex from chicken condensin I revealed by cross-linking and molecular modelling
- PMID: 25716199
- PMCID: PMC4345284
- DOI: 10.1098/rsob.150005
Three-dimensional topology of the SMC2/SMC4 subcomplex from chicken condensin I revealed by cross-linking and molecular modelling
Abstract
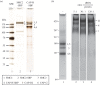
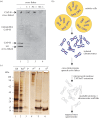
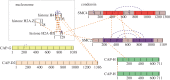
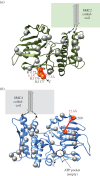
SMC proteins are essential components of three protein complexes that are important for chromosome structure and function. The cohesin complex holds replicated sister chromatids together, whereas the condensin complex has an essential role in mitotic chromosome architecture. Both are involved in interphase genome organization. SMC-containing complexes are large (more than 650 kDa for condensin) and contain long anti-parallel coiled-coils. They are thus difficult subjects for conventional crystallographic and electron cryomicroscopic studies. Here, we have used amino acid-selective cross-linking and mass spectrometry combined with structure prediction to develop a full-length molecular draft three-dimensional structure of the SMC2/SMC4 dimeric backbone of chicken condensin. We assembled homology-based molecular models of the globular heads and hinges with the lengthy coiled-coils modelled in fragments, using numerous high-confidence cross-links and accounting for potential irregularities. Our experiments reveal that isolated condensin complexes can exist with their coiled-coil segments closely apposed to one another along their lengths and define the relative spatial alignment of the two anti-parallel coils. The centres of the coiled-coils can also approach one another closely in situ in mitotic chromosomes. In addition to revealing structural information, our cross-linking data suggest that both H2A and H4 may have roles in condensin interactions with chromatin.
Keywords: SMC; coiled-coil; condensin; cross-linking; mass spectrometry; structure.
Figures




References
-
- Hirano T. 1999. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 13, 11–19 (doi:10.1101/gad.13.1.11) - DOI - PubMed
-
- Nolivos S, Sherratt D. 2014. The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol. Rev. 38, 380–392 (doi:10.1111/1574-6976.12045) - DOI - PMC - PubMed
-
- Jessberger R. 2002. The many functions of SMC proteins in chromosome dynamics. Nat. Rev. Mol. Cell Biol. 3, 767–778 (doi:10.1038/nrm930) - DOI - PubMed
-
- Hudson DF, Marshall KM, Earnshaw WC. 2009. Condensin: architect of mitotic chromosomes. Chromosome Res. 17, 131–144 (doi:10.1007/s10577-008-9009-7) - DOI - PubMed
-
- Wood AJ, Severson AF, Meyer BJ. 2010. Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 11, 391–404 (doi:10.1038/nrg2794) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

