Insights on the emergence of Mycobacterium tuberculosis from the analysis of Mycobacterium kansasii
- PMID: 25716827
- PMCID: PMC5322544
- DOI: 10.1093/gbe/evv035
Insights on the emergence of Mycobacterium tuberculosis from the analysis of Mycobacterium kansasii
Abstract
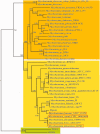
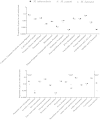
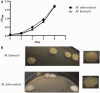
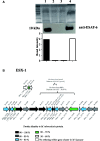
By phylogenetic analysis, Mycobacterium kansasii is closely related to Mycobacterium tuberculosis. Yet, although both organisms cause pulmonary disease, M. tuberculosis is a global health menace, whereas M. kansasii is an opportunistic pathogen. To illuminate the differences between these organisms, we have sequenced the genome of M. kansasii ATCC 12478 and its plasmid (pMK12478) and conducted side-by-side in vitro and in vivo investigations of these two organisms. The M. kansasii genome is 6,432,277 bp, more than 2 Mb longer than that of M. tuberculosis H37Rv, and the plasmid contains 144,951 bp. Pairwise comparisons reveal conserved and discordant genes and genomic regions. A notable example of genomic conservation is the virulence locus ESX-1, which is intact and functional in the low-virulence M. kansasii, potentially mediating phagosomal disruption. Differences between these organisms include a decreased predicted metabolic capacity, an increased proportion of toxin-antitoxin genes, and the acquisition of M. tuberculosis-specific genes in the pathogen since their common ancestor. Consistent with their distinct epidemiologic profiles, following infection of C57BL/6 mice, M. kansasii counts increased by less than 10-fold over 6 weeks, whereas M. tuberculosis counts increased by over 10,000-fold in just 3 weeks. Together, these data suggest that M. kansasii can serve as an image of the environmental ancestor of M. tuberculosis before its emergence as a professional pathogen, and can be used as a model organism to study the switch from an environmental opportunistic pathogen to a professional host-restricted pathogen.
Keywords: Mycobacterium kansasii; comparative genomics; mycobacteria; phylogeny; virulence.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

