Genome editing strategies: potential tools for eradicating HIV-1/AIDS
- PMID: 25716921
- PMCID: PMC4433555
- DOI: 10.1007/s13365-014-0308-9
Genome editing strategies: potential tools for eradicating HIV-1/AIDS
Abstract
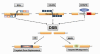
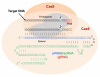
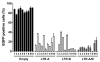
Current therapy for controlling human immunodeficiency virus (HIV-1) infection and preventing acquired immunodeficiency syndrome (AIDS) progression has profoundly decreased viral replication in cells susceptible to HIV-1 infection, but it does not eliminate the low level of viral replication in latently infected cells, which contain integrated copies of HIV-1 proviral DNA. There is an urgent need for the development of HIV-1 genome eradication strategies that will lead to a permanent or "sterile" cure of HIV-1/AIDS. In the past few years, novel nuclease-initiated genome editing tools have been developing rapidly, including zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the CRISPR/Cas9 system. These surgical knives, which can excise any genome, provide a great opportunity to eradicate the HIV-1 genome by targeting highly conserved regions of the HIV-1 long terminal repeats or essential viral genes. Given the time consuming and costly engineering of target-specific ZFNs and TALENs, the RNA-guided endonuclease Cas9 technology has emerged as a simpler and more versatile technology to allow permanent removal of integrated HIV-1 proviral DNA in eukaryotic cells, and hopefully animal models or human patients. The major unmet challenges of this approach at present include inefficient nuclease gene delivery, potential off-target cleavage, and cell-specific genome targeting. Nanoparticle or lentivirus-mediated delivery of next generation Cas9 technologies including nickase or RNA-guided FokI nuclease (RFN) will further improve the potential for genome editing to become a promising approach for curing HIV-1/AIDS.
Figures

References
-
- Allers K, Hutter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, Schneider T. Evidence for the cure of HIV infection by CCR5Delta32/Delta32 stem cell transplantation. Blood. 2011;117:2791–9. - PubMed
-
- Archin NM, Bateson R, Tripathy MK, Crooks AM, Yang KH, Dahl NP, Kearney MF, Anderson EM, Coffin JM, Strain MC, Richman DD, Robertson KR, Kashuba AD, Bosch RJ, Hazuda DJ, Kuruc JD, Eron JJ, Margolis DM. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis. 2014;210:728–35. - PMC - PubMed
-
- Arnould S, Delenda C, Grizot S, Desseaux C, Paques F, Silva GH, Smith J. The I-CreI meganuclease and its engineered derivatives: applications from cell modification to gene therapy. Protein Eng Des Sel. 2011;24:27–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

