Human Papillomavirus 16 Oncoprotein Expression Is Controlled by the Cellular Splicing Factor SRSF2 (SC35)
- PMID: 25717103
- PMCID: PMC4442513
- DOI: 10.1128/JVI.03434-14
Human Papillomavirus 16 Oncoprotein Expression Is Controlled by the Cellular Splicing Factor SRSF2 (SC35)
Abstract
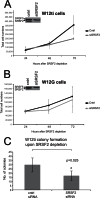
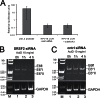
High-risk human papillomaviruses (HR-HPV) cause anogenital cancers, including cervical cancer, and head and neck cancers. Human papillomavirus 16 (HPV16) is the most prevalent HR-HPV. HPV oncogenesis is driven by two viral oncoproteins, E6 and E7, which are expressed through alternative splicing of a polycistronic RNA to yield four major splice isoforms (E6 full length, E6*I, E6*II, E6*X). The production of multiple mRNA isoforms from a single gene is controlled by serine/arginine-rich splicing factors (SRSFs), and HPV16 infection induces overexpression of a subset of these, SRSFs 1, 2, and 3. In this study, we examined whether these proteins could control HPV16 oncoprotein expression. Small interfering RNA (siRNA) depletion experiments revealed that SRSF1 did not affect oncoprotein RNA levels. While SRSF3 knockdown caused some reduction in E6E7 expression, depletion of SRSF2 resulted in a significant loss of E6E7 RNAs, resulting in reduced levels of the E6-regulated p53 proteins and E7 oncoprotein itself. SRSF2 contributed to the tumor phenotype of HPV16-positive cervical cancer cells, as its depletion resulted in decreased cell proliferation, reduced colony formation, and increased apoptosis. SRSF2 did not affect transcription from the P97 promoter that controls viral oncoprotein expression. Rather, RNA decay experiments showed that SRSF2 is required to maintain stability of E6E7 mRNAs. These data show that SRSF2 is a key regulator of HPV16 oncoprotein expression and cervical tumor maintenance.
Importance: Expression of the HPV16 oncoproteins E7 and E6 drives HPV-associated tumor formation. Although increased transcription may yield increased levels of E6E7 mRNAs, it is known that the RNAs can have increased stability upon integration into the host genome. SR splicing factors (SRSFs) control splicing but can also control other events in the RNA life cycle, including RNA stability. Previously, we demonstrated increased levels of SRSFs 1, 2, and 3 during cervical tumor progression. Now we show that SRSF2 is required for expression of E6E7 mRNAs in cervical tumor but not nontumor cells and may act by inhibiting their decay. SRSF2 depletion in W12 tumor cells resulted in increased apoptosis, decreased proliferation, and decreased colony formation, suggesting that SRSF2 has oncogenic functions in cervical tumor progression. SRSF function can be targeted by known drugs that inhibit SRSF phosphorylation, suggesting a possible new avenue in abrogating HPV oncoprotein activity.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures






Similar articles
-
Heterogeneous Nuclear Ribonucleoprotein A1 (hnRNP A1) and hnRNP A2 Inhibit Splicing to Human Papillomavirus 16 Splice Site SA409 through a UAG-Containing Sequence in the E7 Coding Region.J Virol. 2020 Sep 29;94(20):e01509-20. doi: 10.1128/JVI.01509-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32759322 Free PMC article.
-
Human Papillomavirus Type 16 Circular RNA Is Barely Detectable for the Claimed Biological Activity.mBio. 2022 Feb 22;13(1):e0359421. doi: 10.1128/mbio.03594-21. Epub 2022 Jan 18. mBio. 2022. PMID: 35038914 Free PMC article.
-
In vitro and in vivo growth inhibition of human cervical cancer cells via human papillomavirus E6/E7 mRNAs' cleavage by CRISPR/Cas13a system.Antiviral Res. 2020 Jun;178:104794. doi: 10.1016/j.antiviral.2020.104794. Epub 2020 Apr 14. Antiviral Res. 2020. PMID: 32298665
-
High-Risk Human Papillomavirus Oncogenic E6/E7 mRNAs Splicing Regulation.Front Cell Infect Microbiol. 2022 Jun 27;12:929666. doi: 10.3389/fcimb.2022.929666. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35832386 Free PMC article. Review.
-
Alternative splicing in the genome of HPV and its regulation.Front Cell Infect Microbiol. 2024 Oct 22;14:1443868. doi: 10.3389/fcimb.2024.1443868. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39502170 Free PMC article. Review.
Cited by
-
PRMT1 Modulates Alternative Splicing to Enhance HPV18 mRNA Stability and Promote the Establishment of Infection.bioRxiv [Preprint]. 2024 Sep 26:2024.09.26.614592. doi: 10.1101/2024.09.26.614592. bioRxiv. 2024. PMID: 39386465 Free PMC article. Preprint.
-
Physiological and Pathological Function of Serine/Arginine-Rich Splicing Factor 4 and Related Diseases.Biomed Res Int. 2018 Feb 12;2018:3819719. doi: 10.1155/2018/3819719. eCollection 2018. Biomed Res Int. 2018. PMID: 29789787 Free PMC article. Review.
-
Serine/Arginine-Rich Splicing Factor 3 and Heterogeneous Nuclear Ribonucleoprotein A1 Regulate Alternative RNA Splicing and Gene Expression of Human Papillomavirus 18 through Two Functionally Distinguishable cis Elements.J Virol. 2016 Sep 29;90(20):9138-52. doi: 10.1128/JVI.00965-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27489271 Free PMC article.
-
Interplay Between CMGC Kinases Targeting SR Proteins and Viral Replication: Splicing and Beyond.Front Microbiol. 2021 Mar 29;12:658721. doi: 10.3389/fmicb.2021.658721. eCollection 2021. Front Microbiol. 2021. PMID: 33854493 Free PMC article. Review.
-
Serine/Arginine-rich Splicing Factor 2 Modulates Herpes Simplex Virus Type 1 Replication via Regulating Viral Gene Transcriptional Activity and Pre-mRNA Splicing.J Biol Chem. 2016 Dec 16;291(51):26377-26387. doi: 10.1074/jbc.M116.753046. Epub 2016 Oct 26. J Biol Chem. 2016. PMID: 27784784 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

