Metabolic Regulation by Lysine Malonylation, Succinylation, and Glutarylation
- PMID: 25717114
- PMCID: PMC4563717
- DOI: 10.1074/mcp.R114.046664
Metabolic Regulation by Lysine Malonylation, Succinylation, and Glutarylation
Abstract
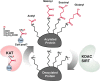
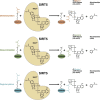
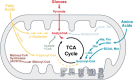
Protein acetylation is a well-studied regulatory mechanism for several cellular processes, ranging from gene expression to metabolism. Recent discoveries of new post-translational modifications, including malonylation, succinylation, and glutarylation, have expanded our understanding of the types of modifications found on proteins. These three acidic lysine modifications are structurally similar but have the potential to regulate different proteins in different pathways. The deacylase sirtuin 5 (SIRT5) catalyzes the removal of these modifications from a wide range of proteins in different subcellular compartments. Here, we review these new modifications, their regulation by SIRT5, and their emerging role in cellular regulation and diseases.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
References
-
- Roth S. Y., Denu J. M., Allis C. D. (2001) Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120 - PubMed
-
- Gu W., Roeder R. G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 - PubMed
-
- Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X. F., Yao T. P. (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30AG028716-01/AG/NIA NIH HHS/United States
- P30 DK096493/DK/NIDDK NIH HHS/United States
- R01AG045351/AG/NIA NIH HHS/United States
- R01AA022146/AA/NIAAA NIH HHS/United States
- R01GM105933/GM/NIGMS NIH HHS/United States
- U54RR020389/RR/NCRR NIH HHS/United States
- U24CA160036/CA/NCI NIH HHS/United States
- R01 AG045351/AG/NIA NIH HHS/United States
- U24 CA160036/CA/NCI NIH HHS/United States
- 5P30DK096493-02/DK/NIDDK NIH HHS/United States
- L40 RR020389/RR/NCRR NIH HHS/United States
- R01 GM105933/GM/NIGMS NIH HHS/United States
- P30 AG028716/AG/NIA NIH HHS/United States
- R01 AA022146/AA/NIAAA NIH HHS/United States
LinkOut - more resources
Full Text Sources

