Automated benchmarking of peptide-MHC class I binding predictions
- PMID: 25717196
- PMCID: PMC4481849
- DOI: 10.1093/bioinformatics/btv123
Automated benchmarking of peptide-MHC class I binding predictions
Abstract
Motivation: Numerous in silico methods predicting peptide binding to major histocompatibility complex (MHC) class I molecules have been developed over the last decades. However, the multitude of available prediction tools makes it non-trivial for the end-user to select which tool to use for a given task. To provide a solid basis on which to compare different prediction tools, we here describe a framework for the automated benchmarking of peptide-MHC class I binding prediction tools. The framework runs weekly benchmarks on data that are newly entered into the Immune Epitope Database (IEDB), giving the public access to frequent, up-to-date performance evaluations of all participating tools. To overcome potential selection bias in the data included in the IEDB, a strategy was implemented that suggests a set of peptides for which different prediction methods give divergent predictions as to their binding capability. Upon experimental binding validation, these peptides entered the benchmark study.
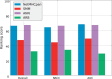
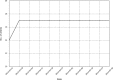
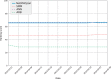
Results: The benchmark has run for 15 weeks and includes evaluation of 44 datasets covering 17 MHC alleles and more than 4000 peptide-MHC binding measurements. Inspection of the results allows the end-user to make educated selections between participating tools. Of the four participating servers, NetMHCpan performed the best, followed by ANN, SMM and finally ARB.
Availability and implementation: Up-to-date performance evaluations of each server can be found online at http://tools.iedb.org/auto_bench/mhci/weekly. All prediction tool developers are invited to participate in the benchmark. Sign-up instructions are available at http://tools.iedb.org/auto_bench/mhci/join.
Contact: mniel@cbs.dtu.dk or bpeters@liai.org
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures



References
-
- Bui H.-H., et al. . (2005) Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics, 57, 304–314. - PubMed
-
- Eyrich V.A., et al. . (2001) EVA: continuous automatic evaluation of protein structure prediction servers. Bioinformatics, 17, 1242–1243. - PubMed
-
- Hattotuwagama C.K., et al. . (2004) Quantitative online prediction of peptide binding to the major histocompatibility complex. J. Mol. Graph. Model.,22, 195–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

