Characterization of interactions between inclusion membrane proteins from Chlamydia trachomatis
- PMID: 25717440
- PMCID: PMC4324299
- DOI: 10.3389/fcimb.2015.00013
Characterization of interactions between inclusion membrane proteins from Chlamydia trachomatis
Abstract
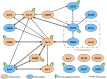
Chlamydiae are obligate intracellular pathogens of eukaryotes. The bacteria grow in an intracellular vesicle called an inclusion, the membrane of which is heavily modified by chlamydial proteins called Incs (Inclusion membrane proteins). Incs represent 7-10% of the genomes of Chlamydia and, given their localization at the interface between the host and the pathogen, likely play a key role in the development and pathogenesis of the bacterium. However, their functions remain largely unknown. Here, we characterized the interaction properties between various Inc proteins of C. trachomatis, using a bacterial two-hybrid (BACTH) method suitable for detecting interactions between integral membrane proteins. To validate this approach, we first examined the oligomerization properties of the well-characterized IncA protein and showed that both the cytoplasmic domain and the transmembrane region independently contribute to IncA oligomerization. We then analyzed a set of Inc proteins and identified novel interactions between these components. Two small Incs, IncF, and Ct222, were found here to interact with many other Inc proteins and may thus represent interaction nodes within the inclusion membrane. Our data suggest that the Inc proteins may assemble in the membrane of the inclusion to form specific multi-molecular complexes in an hierarchical and temporal manner. These studies will help to better define the putative functions of the Inc proteins in the infectious process of Chlamydia.
Keywords: BACTH; Chlamydia; Inc protein; bacterial two-hybrid system; inclusion membrane proteins; protein-protein interactions.
Figures




References
-
- Agaisse H., Derré I. (2014). Expression of the effector protein IncD in Chlamydia trachomatis mediates recruitment of the lipid transfer protein CERT and the endoplasmic reticulum-resident protein VAPB to the inclusion membrane. Infect. Immun. 82, 2037–2047. 10.1128/IAI.01530-14 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

