Thiol-based redox switches in prokaryotes
- PMID: 25720121
- PMCID: PMC4438307
- DOI: 10.1515/hsz-2015-0102
Thiol-based redox switches in prokaryotes
Abstract
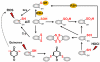
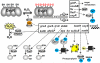
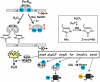
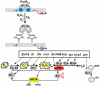
Bacteria encounter reactive oxygen species (ROS) as a consequence of the aerobic life or as an oxidative burst of activated neutrophils during infections. In addition, bacteria are exposed to other redox-active compounds, including hypochloric acid (HOCl) and reactive electrophilic species (RES) such as quinones and aldehydes. These reactive species often target the thiol groups of cysteines in proteins and lead to thiol-disulfide switches in redox-sensing regulators to activate specific detoxification pathways and to restore the redox balance. Here, we review bacterial thiol-based redox sensors that specifically sense ROS, RES and HOCl via thiol-based mechanisms and regulate gene transcription in Gram-positive model bacteria and in human pathogens, such as Staphylococcus aureus and Mycobacterium tuberculosis. We also pay particular attention to emerging widely conserved HOCl-specific redox regulators that have been recently characterized in Escherichia coli. Different mechanisms are used to sense and respond to ROS, RES and HOCl by 1-Cys-type and 2-Cys-type thiol-based redox sensors that include versatile thiol-disulfide switches (OxyR, OhrR, HypR, YodB, NemR, RclR, Spx, RsrA/RshA) or alternative Cys phosphorylations (SarZ, MgrA, SarA), thiol-S-alkylation (QsrR), His-oxidation (PerR) and methionine oxidation (HypT). In pathogenic bacteria, these redox-sensing regulators are often important virulence regulators and required for adapation to the host immune defense.
Figures

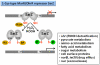
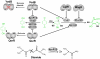
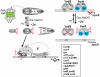

Similar articles
-
Two distinct mechanisms of transcriptional regulation by the redox sensor YodB.Proc Natl Acad Sci U S A. 2016 Aug 30;113(35):E5202-11. doi: 10.1073/pnas.1604427113. Epub 2016 Aug 16. Proc Natl Acad Sci U S A. 2016. PMID: 27531959 Free PMC article.
-
Thiol-based redox switches and gene regulation.Antioxid Redox Signal. 2011 Mar 15;14(6):1049-63. doi: 10.1089/ars.2010.3400. Epub 2010 Oct 28. Antioxid Redox Signal. 2011. PMID: 20626317 Free PMC article. Review.
-
The MarR/DUF24-Family QsrR Repressor Senses Quinones and Oxidants by Thiol Switch Mechanisms in Staphylococcus aureus.Antioxid Redox Signal. 2023 May;38(13-15):877-895. doi: 10.1089/ars.2022.0090. Epub 2022 Nov 28. Antioxid Redox Signal. 2023. PMID: 36242097
-
Thiol-based redox switches in the major pathogen Staphylococcus aureus.Biol Chem. 2020 Nov 23;402(3):333-361. doi: 10.1515/hsz-2020-0272. Print 2021 Feb 23. Biol Chem. 2020. PMID: 33544504 Review.
-
Redox active thiol sensors of oxidative and nitrosative stress.Antioxid Redox Signal. 2012 Nov 1;17(9):1201-14. doi: 10.1089/ars.2012.4522. Epub 2012 Mar 15. Antioxid Redox Signal. 2012. PMID: 22257022 Free PMC article. Review.
Cited by
-
Two distinct mechanisms of transcriptional regulation by the redox sensor YodB.Proc Natl Acad Sci U S A. 2016 Aug 30;113(35):E5202-11. doi: 10.1073/pnas.1604427113. Epub 2016 Aug 16. Proc Natl Acad Sci U S A. 2016. PMID: 27531959 Free PMC article.
-
The Catalase KatA Contributes to Microaerophilic H2O2 Priming to Acquire an Improved Oxidative Stress Resistance in Staphylococcus aureus.Antioxidants (Basel). 2022 Sep 12;11(9):1793. doi: 10.3390/antiox11091793. Antioxidants (Basel). 2022. PMID: 36139867 Free PMC article.
-
Mycobacterial DnaB helicase intein as oxidative stress sensor.Nat Commun. 2018 Oct 19;9(1):4363. doi: 10.1038/s41467-018-06554-x. Nat Commun. 2018. PMID: 30341292 Free PMC article.
-
OsnR is an autoregulatory negative transcription factor controlling redox-dependent stress responses in Corynebacterium glutamicum.Microb Cell Fact. 2021 Oct 18;20(1):203. doi: 10.1186/s12934-021-01693-1. Microb Cell Fact. 2021. PMID: 34663317 Free PMC article.
-
Ohr and OhrR Are Critical for Organic Peroxide Resistance and Symbiosis in Azorhizobium caulinodans ORS571.Genes (Basel). 2020 Mar 20;11(3):335. doi: 10.3390/genes11030335. Genes (Basel). 2020. PMID: 32245101 Free PMC article.
References
-
- Antelmann H, Hecker M, Zuber P. Proteomic signatures uncover thiol-specific electrophile resistance mechanisms in Bacillus subtilis. Expert Rev Proteomics. 2008;5:77–90. - PubMed
-
- Atichartpongkul S, Loprasert S, Vattanaviboon P, Whangsuk W, Helmann JD, Mongkolsuk S. Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology. 2001;147:1775–1782. - PubMed
-
- Bae JB, Park JH, Hahn MY, Kim MS, Roe JH. Redox-dependent changes in RsrA, an anti-sigma factor in Streptomyces coelicolor: zinc release and disulfide bond formation. J Mol Biol. 2004;335:425–435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
