Studying the dynamics of coronavirus replicative structures
- PMID: 25720487
- PMCID: PMC7122990
- DOI: 10.1007/978-1-4939-2438-7_22
Studying the dynamics of coronavirus replicative structures
Abstract
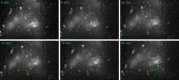
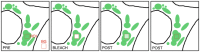
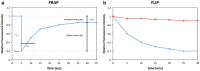
Coronaviruses (CoVs) generate specialized membrane compartments, which consist of double membrane vesicles connected to convoluted membranes, the so-called replicative structures, where viral RNA synthesis takes place. These sites harbor the CoV replication-transcription complexes (RTCs): multi-protein complexes consisting of 16 nonstructural proteins (nsps), the CoV nucleocapsid protein (N) and presumably host proteins. To successfully establish functional membrane-bound RTCs all of the viral and host constituents need to be correctly spatiotemporally organized during viral infection. Few studies, however, have investigated the dynamic processes involved in the formation and functioning of the (subunits of) CoV RTCs and the replicative structures in living cells. In this chapter we describe several protocols to perform time-lapse imaging of CoV-infected cells and to study the kinetics of (subunits of) the CoV replicative structures. The approaches described are not limited to CoV-infected cells; they can also be applied to other virus-infected or non-infected cells.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

