3D hotspots of recurrent retroviral insertions reveal long-range interactions with cancer genes
- PMID: 25721899
- PMCID: PMC4351571
- DOI: 10.1038/ncomms7381
3D hotspots of recurrent retroviral insertions reveal long-range interactions with cancer genes
Abstract
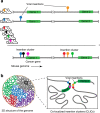
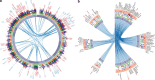
Genomically distal mutations can contribute to the deregulation of cancer genes by engaging in chromatin interactions. To study this, we overlay viral cancer-causing insertions obtained in a murine retroviral insertional mutagenesis screen with genome-wide chromatin conformation capture data. Here we find that insertions tend to cluster in 3D hotspots within the nucleus. The identified hotspots are significantly enriched for known cancer genes, and bear the expected characteristics of bona fide regulatory interactions, such as enrichment for transcription factor-binding sites. In addition, we observe a striking pattern of mutual exclusive integration. This is an indication that insertions in these loci target the same gene, either in their linear genomic vicinity or in their 3D spatial vicinity. Our findings shed new light on the repertoire of targets obtained from insertional mutagenesis screening and underline the importance of considering the genome as a 3D structure when studying effects of genomic perturbations.
Conflict of interest statement
There are no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

