Second-generation PLINK: rising to the challenge of larger and richer datasets
- PMID: 25722852
- PMCID: PMC4342193
- DOI: 10.1186/s13742-015-0047-8
Second-generation PLINK: rising to the challenge of larger and richer datasets
Abstract
Background: PLINK 1 is a widely used open-source C/C++ toolset for genome-wide association studies (GWAS) and research in population genetics. However, the steady accumulation of data from imputation and whole-genome sequencing studies has exposed a strong need for faster and scalable implementations of key functions, such as logistic regression, linkage disequilibrium estimation, and genomic distance evaluation. In addition, GWAS and population-genetic data now frequently contain genotype likelihoods, phase information, and/or multiallelic variants, none of which can be represented by PLINK 1's primary data format.
Findings: To address these issues, we are developing a second-generation codebase for PLINK. The first major release from this codebase, PLINK 1.9, introduces extensive use of bit-level parallelism, [Formula: see text]-time/constant-space Hardy-Weinberg equilibrium and Fisher's exact tests, and many other algorithmic improvements. In combination, these changes accelerate most operations by 1-4 orders of magnitude, and allow the program to handle datasets too large to fit in RAM. We have also developed an extension to the data format which adds low-overhead support for genotype likelihoods, phase, multiallelic variants, and reference vs. alternate alleles, which is the basis of our planned second release (PLINK 2.0).
Conclusions: The second-generation versions of PLINK will offer dramatic improvements in performance and compatibility. For the first time, users without access to high-end computing resources can perform several essential analyses of the feature-rich and very large genetic datasets coming into use.
Keywords: Computational statistics; GWAS; High-density SNP genotyping; Population genetics; Whole-genome sequencing.
Figures
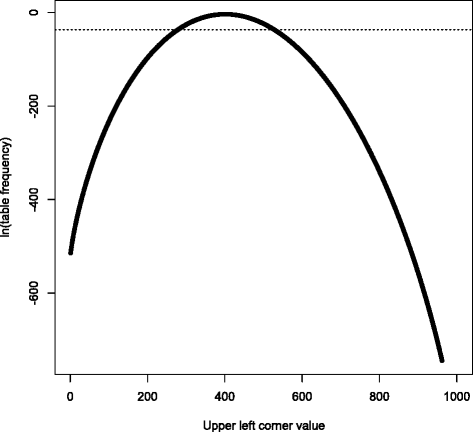
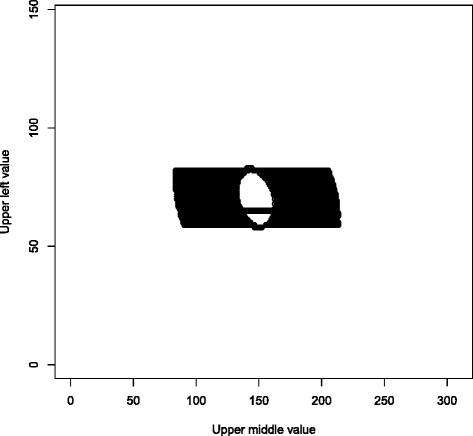
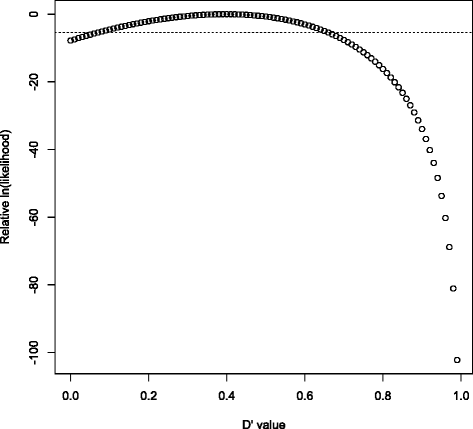
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

