Gateways to the FANTOM5 promoter level mammalian expression atlas
- PMID: 25723102
- PMCID: PMC4310165
- DOI: 10.1186/s13059-014-0560-6
Gateways to the FANTOM5 promoter level mammalian expression atlas
Abstract
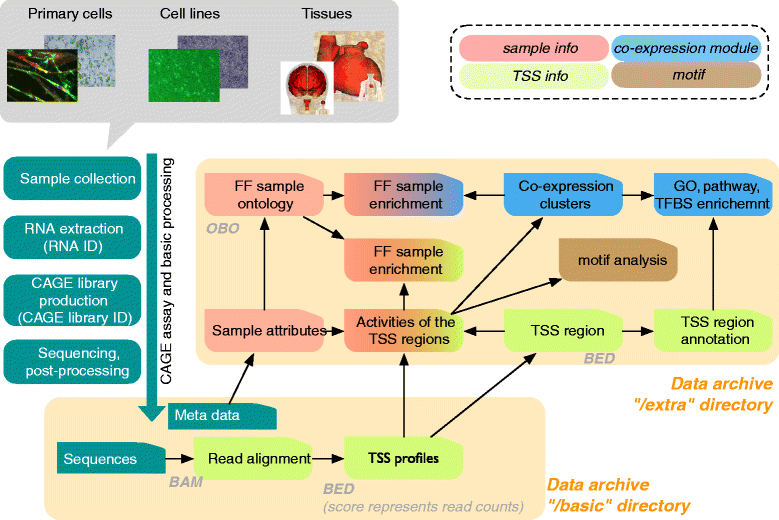
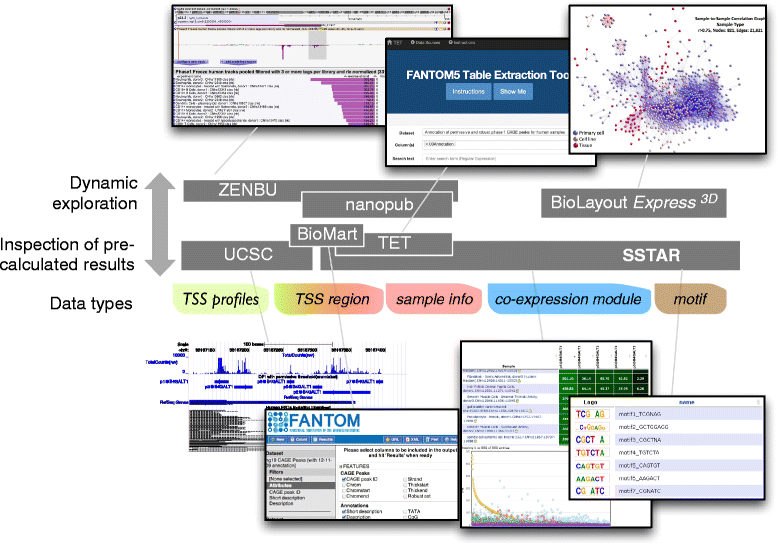
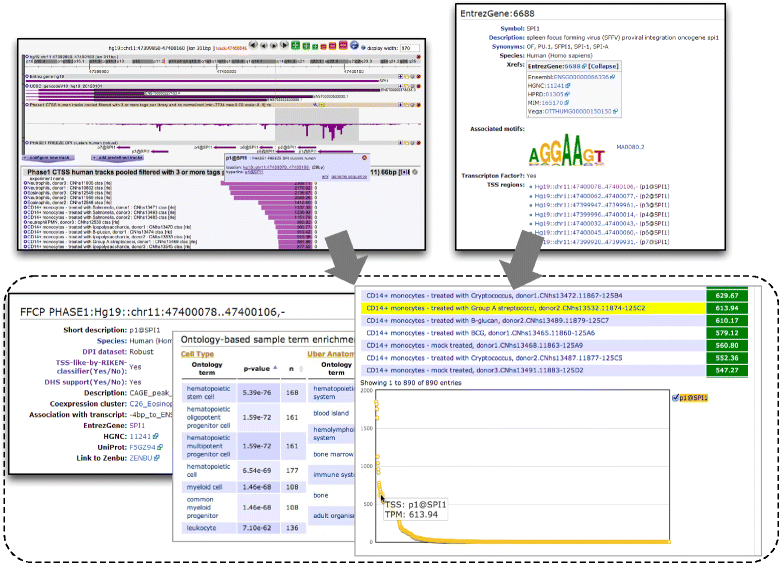
The FANTOM5 project investigates transcription initiation activities in more than 1,000 human and mouse primary cells, cell lines and tissues using CAGE. Based on manual curation of sample information and development of an ontology for sample classification, we assemble the resulting data into a centralized data resource (http://fantom.gsc.riken.jp/5/). This resource contains web-based tools and data-access points for the research community to search and extract data related to samples, genes, promoter activities, transcription factors and enhancers across the FANTOM5 atlas.
Figures

References
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science 2005, 309:1559–1563. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

