Structural basis for Marburg virus neutralization by a cross-reactive human antibody
- PMID: 25723165
- PMCID: PMC4344967
- DOI: 10.1016/j.cell.2015.01.041
Structural basis for Marburg virus neutralization by a cross-reactive human antibody
Abstract
The filoviruses, including Marburg and Ebola, express a single glycoprotein on their surface, termed GP, which is responsible for attachment and entry of target cells. Filovirus GPs differ by up to 70% in protein sequence, and no antibodies are yet described that cross-react among them. Here, we present the 3.6 Å crystal structure of Marburg virus GP in complex with a cross-reactive antibody from a human survivor, and a lower resolution structure of the antibody bound to Ebola virus GP. The antibody, MR78, recognizes a GP1 epitope conserved across the filovirus family, which likely represents the binding site of their NPC1 receptor. Indeed, MR78 blocks binding of the essential NPC1 domain C. These structures and additional small-angle X-ray scattering of mucin-containing MARV and EBOV GPs suggest why such antibodies were not previously elicited in studies of Ebola virus, and provide critical templates for development of immunotherapeutics and inhibitors of entry.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


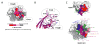


References
-
- Albarino CG, Shoemaker T, Khristova ML, Wamala JF, Muyembe JJ, Balinandi S, Tumusiime A, Campbell S, Cannon D, Gibbons A, et al. Genomic analysis of filoviruses associated with four viral hemorrhagic fever outbreaks in Uganda and the Democratic Republic of the Congo in 2012. Virology. 2013;442:97–100. - PMC - PubMed
-
- Barbey-Martin C, Gigant B, Bizebard T, Calder LJ, Wharton SA, Skehel JJ, Knossow M. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology. 2002;294:70–74. - PubMed
-
- Barrette RW, Metwally SA, Rowland JM, Xu L, Zaki SR, Nichol ST, Rollin PE, Towner JS, Shieh WJ, Batten B, et al. Discovery of swine as a host for the Reston ebolavirus. Science. 2009;325:204–206. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

