Bridger: a new framework for de novo transcriptome assembly using RNA-seq data
- PMID: 25723335
- PMCID: PMC4342890
- DOI: 10.1186/s13059-015-0596-2
Bridger: a new framework for de novo transcriptome assembly using RNA-seq data
Abstract
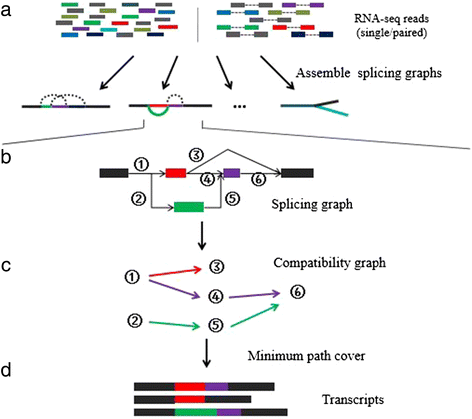
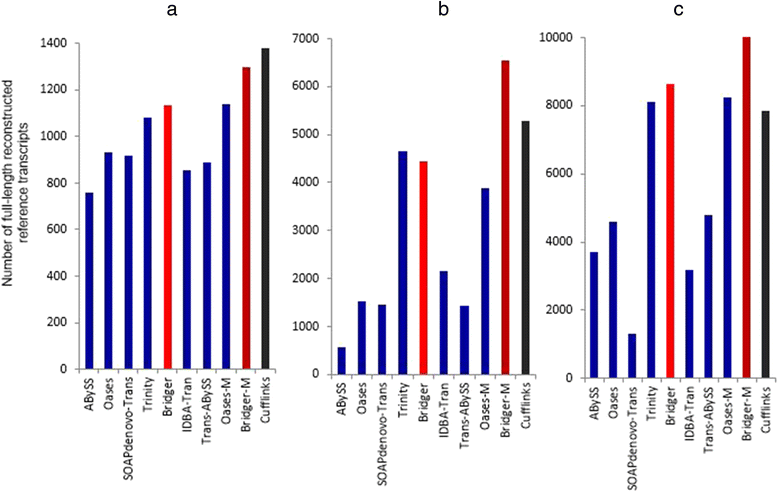
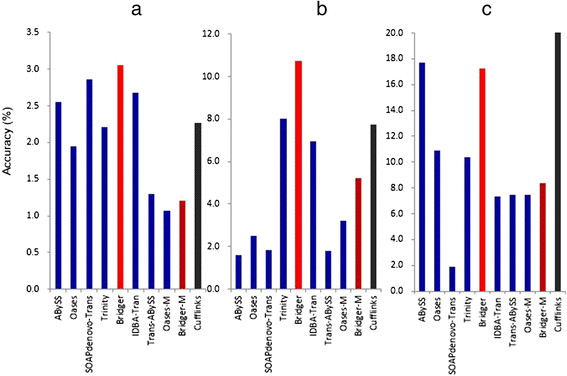
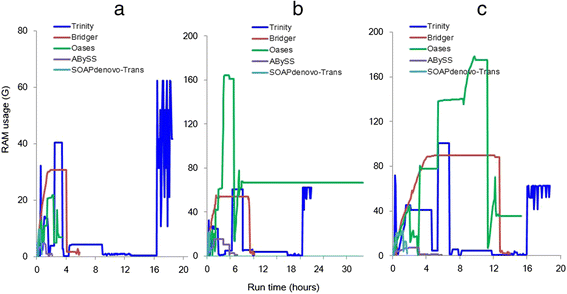
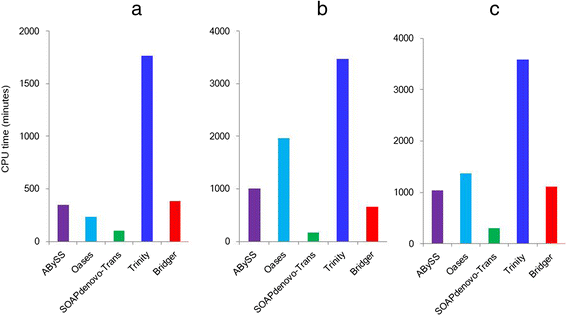
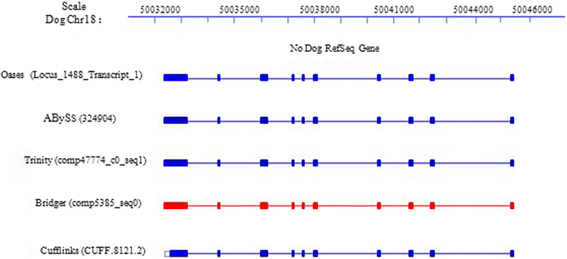
We present a new de novo transcriptome assembler, Bridger, which takes advantage of techniques employed in Cufflinks to overcome limitations of the existing de novo assemblers. When tested on dog, human, and mouse RNA-seq data, Bridger assembled more full-length reference transcripts while reporting considerably fewer candidate transcripts, hence greatly reducing false positive transcripts in comparison with the state-of-the-art assemblers. It runs substantially faster and requires much less memory space than most assemblers. More interestingly, Bridger reaches a comparable level of sensitivity and accuracy with Cufflinks. Bridger is available at https://sourceforge.net/projects/rnaseqassembly/files/?source=navbar.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

