N6-adenosine methylation in MiRNAs
- PMID: 25723394
- PMCID: PMC4344304
- DOI: 10.1371/journal.pone.0118438
N6-adenosine methylation in MiRNAs
Abstract
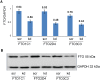
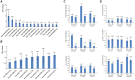
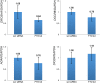
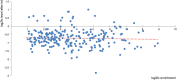
Methylation of N6-adenosine (m6A) has been observed in many different classes of RNA, but its prevalence in microRNAs (miRNAs) has not yet been studied. Here we show that a knockdown of the m6A demethylase FTO affects the steady-state levels of several miRNAs. Moreover, RNA immunoprecipitation with an anti-m6A-antibody followed by RNA-seq revealed that a significant fraction of miRNAs contains m6A. By motif searches we have discovered consensus sequences discriminating between methylated and unmethylated miRNAs. The epigenetic modification of an epigenetic modifier as described here adds a new layer to the complexity of the posttranscriptional regulation of gene expression.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

