The structural biology of CRISPR-Cas systems
- PMID: 25723899
- PMCID: PMC4417044
- DOI: 10.1016/j.sbi.2015.02.002
The structural biology of CRISPR-Cas systems
Abstract
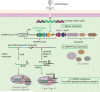
Prokaryotic CRISPR-Cas genomic loci encode RNA-mediated adaptive immune systems that bear some functional similarities with eukaryotic RNA interference. Acquired and heritable immunity against bacteriophage and plasmids begins with integration of ∼30 base pair foreign DNA sequences into the host genome. CRISPR-derived transcripts assemble with CRISPR-associated (Cas) proteins to target complementary nucleic acids for degradation. Here we review recent advances in the structural biology of these targeting complexes, with a focus on structural studies of the multisubunit Type I CRISPR RNA-guided surveillance and the Cas9 DNA endonuclease found in Type II CRISPR-Cas systems. These complexes have distinct structures that are each capable of site-specific double-stranded DNA binding and local helix unwinding.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures

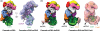


References
-
- Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–339. - PubMed
-
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

