A single epidermal stem cell strategy for safe ex vivo gene therapy
- PMID: 25724200
- PMCID: PMC4403041
- DOI: 10.15252/emmm.201404353
A single epidermal stem cell strategy for safe ex vivo gene therapy
Abstract
There is a widespread agreement from patient and professional organisations alike that the safety of stem cell therapeutics is of paramount importance, particularly for ex vivo autologous gene therapy. Yet current technology makes it difficult to thoroughly evaluate the behaviour of genetically corrected stem cells before they are transplanted. To address this, we have developed a strategy that permits transplantation of a clonal population of genetically corrected autologous stem cells that meet stringent selection criteria and the principle of precaution. As a proof of concept, we have stably transduced epidermal stem cells (holoclones) obtained from a patient suffering from recessive dystrophic epidermolysis bullosa. Holoclones were infected with self-inactivating retroviruses bearing a COL7A1 cDNA and cloned before the progeny of individual stem cells were characterised using a number of criteria. Clonal analysis revealed a great deal of heterogeneity among transduced stem cells in their capacity to produce functional type VII collagen (COLVII). Selected transduced stem cells transplanted onto immunodeficient mice regenerated a non-blistering epidermis for months and produced a functional COLVII. Safety was assessed by determining the sites of proviral integration, rearrangements and hit genes and by whole-genome sequencing. The progeny of the selected stem cells also had a diploid karyotype, was not tumorigenic and did not disseminate after long-term transplantation onto immunodeficient mice. In conclusion, a clonal strategy is a powerful and efficient means of by-passing the heterogeneity of a transduced stem cell population. It guarantees a safe and homogenous medicinal product, fulfilling the principle of precaution and the requirements of regulatory affairs. Furthermore, a clonal strategy makes it possible to envision exciting gene-editing technologies like zinc finger nucleases, TALENs and homologous recombination for next-generation gene therapy.
Keywords: cell therapy; regulatory affairs; stem cells; wound healing.
© 2015 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
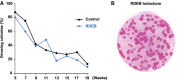
Keratinocytes were isolated from the skin of a 4-year-old patient with severe-generalised RDEB linked to homozygous insertion–deletion in COL7A1 (Hilal et al, 1993). Cultured RDEB cells (blue line) were serially passaged for more than 4 months, displaying a growth potential similar to non-diseased control cells (YF29) isolated from the foreskin of a newborn (black line). To calculate the percentage of growing colonies, 100 to 1,000 cells were plated into indicator dishes at each passage. Cells were grown for 12 days, fixed and stained with rhodamine B. Colonies were scored as growing or aborted (Barrandon & Green, 1987).
Clonal analysis demonstrated the presence of stem cells (holoclones) in a passage VII RDEB culture (95% of growing colonies).

COLVII detection in clones by immunostaining. COLVII expression (green) was detectable in some clones (6, 17, 22, 58 and 61) and not in others (3, 24 and 54); nuclei were stained with Hoechst 33342 (blue). Dotted lines delimit the periphery of keratinocyte colonies from the surrounding irradiated 3T3-J2 feeder cells. Scale bar: 50 μm.
Quantitative RT–PCR analysis of COL7A1 expression in transduced clones compared to untransduced RDEB keratinocytes. All clones shown in (A) were transduced but expressed different levels of COL7A1 transcripts. Clones 6, 17, 22, 54, 58 and 61 expressed higher levels of COL7A1 than control RDEB cells and keratinocytes obtained from healthy donors (YF29 and OR-CA, control 1 and 2, respectively). The level of COL7A1 expression in the RDEB untransduced cells was referenced as 1.
Determination of proviral rearrangements in transduced clones. A Southern blot was performed using genomic DNA of RDEB cells, clones and the infected mass culture from which the clones were isolated. Genomic DNA was digested with EcoRV and SpeI that cut at the 3′ and 5′ end of the provirus (Supplementary Fig S2) and hybridised with a 907-bp COL7A1 probe radiolabelled with 32P isotope. The upper band corresponded to the endogenous signal. The retroviral producer line Flp293A-E1aColVII1 was used as a control for the digested 9.6-kb provirus (proviral signal). Smaller bands corresponded to rearranged proviruses marked with an asterisk.
Identification of stem cells producing COLVII. Western blotting revealed that only clone 6 secreted COLVII in the culture supernatant, while clone 54 and surprisingly clone 22 did not (see A). RDEB cells were used as a negative control and healthy donor cells as a positive control. The secreted matrix metalloproteinase 2 (MMP2) was used as a loading control.

Serial cultivation of transduced and untransduced holoclones demonstrated that the growth potential of the stem cells was not affected by the production of COLVII. Non-COLVII-producing holoclone 54 (black lines) and COLVII-producing holoclone 6 (red lines) were serially transferred once a week until exhaustion (Rochat et al, 1994).
Theoretical number of epidermal cells available for characterisation and CEA production from corrected (clone 6) and uncorrected stem cells (clone 54) calculated from the day of cloning. The colony-forming efficiency and the percentage of growing colonies for each passage were used to calculate the population doubling, the generation number and the total progeny of isolated stem cells. Both corrected and uncorrected epidermal stem cells show high growth potential in vitro.
Immunodeficient SCID mice were transplanted with untransduced cultured RDEB keratinocytes (left) or the COLVII-secreting holoclone (clone 6) (right). Punch biopsies were obtained at various times post-transplantation (PT), stained with haematoxylin/eosine (H/E), for human leucocyte antigen-1 (HLA-1) (green) and human COLVII (red). RDEB keratinocytes generated a HLA-1-positive epidermis that adhered poorly to the dermo-epidermal junction (DEJ) (arrow indicates a blister) and absence of COLVII (dotted line delimits the dermis), whereas the corrected keratinocytes produced an epidermis that adhered to the dermis and deposited COLVII (red) at the DEJ. Note the presence of KI67-positive keratinocytes (red) in the basal and suprabasal layers in the corrected epidermis at 385 days post-transplantation, indicating that transplanted cells had self-renewed for more than a year. Scale bar: 50 μm.
Transmission electron microscopy (TEM) demonstrated the presence of anchoring fibrils (arrows) at the DEJ of the corrected epidermis (middle right panel) and in a normal human skin biopsy (left panel). Note the absence of anchoring fibrils in the RDEB epidermis (middle left panel). Detection of human COLVII by immunogold staining in the DEJ of corrected epidermis (right panel). Scale bar: 250 nm.

Visualisation of COL7A1 sequence by FISH analysis in corrected stem cells at passage XII (clone 6). Five specific signals (red) were detected on five different chromosomes. As expected, a COL7A1 probe (red) hybridised to the two endogenous COL7A1 alleles located on the chromosomes 3 as identified by a specific centromeric probe (Cen 3) and to three other chromosomes identified as chromosomes 2, 11 and 22 by means of specific centromeric probes (Cen 2, Cen 11 and Cen 22, respectively). The latter corresponded to proviruses.
Determination of proviral integration sites in corrected stem cell. Genomic DNA from clone 6 was submitted to whole-genome sequencing together with PCR and subsequent Sanger sequencing to identify the integration sites to the base-pair level. Mate pairs that span from the viral sequence to a human chromosome were extracted and used to estimate integration regions. The reads were mapped to the hg19 reference sequence. Three integration sites were uncovered: one in chromosome 2, one in chromosome 11 and one in chromosome 22, and targeted genes were identified.
Analysis of the level of expression of targeted genes in corrected stem cells by quantitative RT–PCR. DARS was not significantly changed in clone 6 compared to the cells before transduction. Primers used annealed downstream of the proviral integration site. Error bars represent the standard deviation of three replicates.
Identification of sequence abnormalities in the two alleles of the DARS-targeted gene in clone 6 by NGS. Reads were mapped to the hg19 reference sequence. Small insertion–deletions and SNP calling were performed and compared to GWAS databases. The mapping highlighted two indels and one SNP in the DARS genomic sequence. The indels were not associated with disease and the SNP was synonymous (CCU–CCG both codons correspond to proline).

Upper panel: expression of proteins associated with the acquisition of immortalisation process involved in senescence, in evasion of growth suppression and in apoptosis (Hanahan & Weinberg, 2011). The level of P53, P21, RAS and P16 was similar in clone 6, untransduced cells and mass culture from four independent infections, significantly different from the squamous cell carcinoma cell line SCC-13. Lower panel: the phosphorylation state of the PRB restriction point was maintained in clone 6 and untransduced recessive dystrophic epidermolysis bullosa (RDEB) cells, whereas PRB was heavily phosphorylated in transformed cells (SCC-13). Appropriate loading controls were used for each cellular extract (GAPDH for cytoplasmic extracts, histone H3 for nuclear extracts and tubulin for whole-cell extracts. H3 for histone H3, ppRB for hyperphosphorylated pRB and pRB for hypophosphorylated PRB).
Transduced clone 6 had a diploid karyotype at passage XVI (see also Fig5A).
Transduced clones were not tumorigenic. The tumorigenic potential of the clones was tested by tumour formation in nude mice. Transduced RDEB holoclones (blue) [clone 6 (square, n = 4) passage X, clone 22 (circle, n = 2) passage XI, clone 54 (cross, n = 4) passage X] were not tumorigenic when injected subcutaneously in athymic mice as were untransduced RDEB keratinocytes (passage VII). SCC-13, a squamous cell carcinoma cell line (black) (lozenge, n = 2), was used as a positive control.
To test whether the transduced RDEB keratinocytes had disseminated after the generation of an epidermis onto SCID mice (see Fig4C), the internal organs of the recipient mice (3 mice for clone 6 and 2 mice for clone 22) were harvested 385 days post-transplantation and analysed for COL7A1 proviral sequences. No COL7A1 sequence was detected. A mouse transplanted with a holoclone obtained from a healthy donor (YF29) was used as an internal control. PCR-positive controls were genomic DNA from transduced cells (holoclone 6) and cDNAs isolated from healthy keratinocytes. B-actin was run as a loading control.
Comment in
-
Single stem cell gene therapy for genetic skin disease.EMBO Mol Med. 2015 Apr;7(4):366-7. doi: 10.15252/emmm.201404859. EMBO Mol Med. 2015. PMID: 25724199 Free PMC article.
References
-
- Abbott A. Stem-cell ruling riles researchers. Nature. 2013;495:418–419. - PubMed
-
- Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L, Scaramuzza S, Andolfi G, Mirolo M, Brigida I, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360:447–458. - PubMed
-
- Almarza D, Bussadori G, Navarro M, Mavilio F, Larcher F, Murillas R. Risk assessment in skin gene therapy: viral-cellular fusion transcripts generated by proviral transcriptional read-through in keratinocytes transduced with self-inactivating lentiviral vectors. Gene Ther. 2011;18:674–681. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

