Analysis of the history and spread of HIV-1 in Uganda using phylodynamics
- PMID: 25724670
- PMCID: PMC4635457
- DOI: 10.1099/vir.0.000107
Analysis of the history and spread of HIV-1 in Uganda using phylodynamics
Abstract
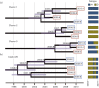
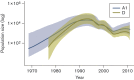
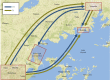
HIV prevalence has decreased in Uganda since the 1990s, but remains substantial within high-risk groups. Here, we reconstruct the history and spread of HIV subtypes A1 and D in Uganda and explore the transmission dynamics in high-risk populations. We analysed HIV pol sequences from female sex workers in Kampala (n = 42), Lake Victoria fisher-folk (n = 46) and a rural clinical cohort (n = 74), together with publicly available sequences from adjacent regions in Uganda (n = 412) and newly generated sequences from samples taken in Kampala in 1986 (n = 12). Of the sequences from the three Ugandan populations, 60 (37.1 %) were classified as subtype D, 54 (33.3 %) as subtype A1, 31 (19.1 %) as A1/D recombinants, six (3.7 %) as subtype C, one (0.6 %) as subtype G and 10 (6.2 %) as other recombinants. Among the A1/D recombinants we identified a new candidate circulating recombinant form. Phylodynamic and phylogeographic analyses using BEAST indicated that the Ugandan epidemics originated in 1960 (1950-1968) for subtype A1 and 1973 (1970-1977) for D, in rural south-western Uganda with subsequent spread to Kampala. They also showed extensive interconnection with adjacent countries. The sequence analysis shows both epidemics grew exponentially during the 1970s-1980s and decreased from 1992, which agrees with HIV prevalence reports in Uganda. Inclusion of sequences from the 1980s indicated the origin of both epidemics was more recent than expected and substantially narrowed the confidence intervals in comparison to previous estimates. We identified three transmission clusters and ten pairs, none of them including patients from different populations, suggesting active transmission within a structured transmission network.
Figures
References
-
- Asiki G., Mpendo J., Abaasa A., Agaba C., Nanvubya A., Nielsen L., Seeley J., Kaleebu P., Grosskurth H., Kamali A. ( 2011. ). HIV and syphilis prevalence and associated risk factors among fishing communities of Lake Victoria, Uganda. Sex Transm Infect 87, 511–515. 10.1136/sti.2010.046805 - DOI - PubMed
-
- Baeten J. M., Chohan B., Lavreys L., Chohan V., McClelland R. S., Certain L., Mandaliya K., Jaoko W., Overbaugh J. ( 2007. ). HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis 195, 1177–1180. 10.1086/512682 - DOI - PubMed
-
- Carswell J. W. ( 1987. ). HIV infection in healthy persons in Uganda. AIDS 1, 223–227. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

