Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface
- PMID: 25725012
- PMCID: PMC4462956
- DOI: 10.1093/femsre/fuu012
Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface
Abstract
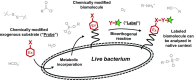
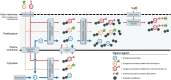
The cell surface is the essential interface between a bacterium and its surroundings. Composed primarily of molecules that are not directly genetically encoded, this highly dynamic structure accommodates the basic cellular processes of growth and division as well as the transport of molecules between the cytoplasm and the extracellular milieu. In this review, we describe aspects of bacterial growth, division and secretion that have recently been uncovered by metabolic labeling of the cell envelope. Metabolite derivatives can be used to label a variety of macromolecules, from proteins to non-genetically-encoded glycans and lipids. The embedded metabolite enables precise tracking in time and space, and the versatility of newer chemoselective detection methods offers the ability to execute multiple experiments concurrently. In addition to reviewing the discoveries enabled by metabolic labeling of the bacterial cell envelope, we also discuss the potential of these techniques for translational applications. Finally, we offer some guidelines for implementing this emerging technology.
Keywords: bioorthogonal; click chemistry; glycolipid; metabolic labeling; peptidoglycan; protein secretion.
© FEMS 2015. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc. 2004;126:15046–7. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

