Automatic concept recognition using the human phenotype ontology reference and test suite corpora
- PMID: 25725061
- PMCID: PMC4343077
- DOI: 10.1093/database/bav005
Automatic concept recognition using the human phenotype ontology reference and test suite corpora
Abstract
Concept recognition tools rely on the availability of textual corpora to assess their performance and enable the identification of areas for improvement. Typically, corpora are developed for specific purposes, such as gene name recognition. Gene and protein name identification are longstanding goals of biomedical text mining, and therefore a number of different corpora exist. However, phenotypes only recently became an entity of interest for specialized concept recognition systems, and hardly any annotated text is available for performance testing and training. Here, we present a unique corpus, capturing text spans from 228 abstracts manually annotated with Human Phenotype Ontology (HPO) concepts and harmonized by three curators, which can be used as a reference standard for free text annotation of human phenotypes. Furthermore, we developed a test suite for standardized concept recognition error analysis, incorporating 32 different types of test cases corresponding to 2164 HPO concepts. Finally, three established phenotype concept recognizers (NCBO Annotator, OBO Annotator and Bio-LarK CR) were comprehensively evaluated, and results are reported against both the text corpus and the test suites. The gold standard and test suites corpora are available from http://bio-lark.org/hpo_res.html. Database URL: http://bio-lark.org/hpo_res.html.
© The Author(s) 2015. Published by Oxford University Press.
Figures

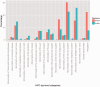
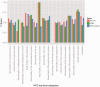
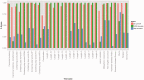
References
-
- Köhler S., Doelken S.C., Ruef B.J., et al. . (2014) Construction and accessibility of a cross-species phenotype ontology along with gene annotations for biomedical research. F1000Research, 2, 30, [v2; ref status: indexed, http://f1000r.es/2td]. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

