Cutting edge: An in vivo reporter reveals active B cell receptor signaling in the germinal center
- PMID: 25725108
- PMCID: PMC4369439
- DOI: 10.4049/jimmunol.1403086
Cutting edge: An in vivo reporter reveals active B cell receptor signaling in the germinal center
Abstract
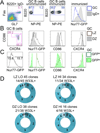
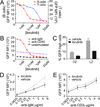
Long-lasting Ab responses rely on the germinal center (GC), where B cells bearing high-affinity Ag receptors are selected from a randomly mutated pool to populate the memory and plasma cell compartments. Signaling downstream of the BCR is dampened in GC B cells, raising the possibility that Ag presentation and competition for T cell help, rather than Ag-dependent signaling per se, drive these critical selection events. In this study we use an in vivo reporter of BCR signaling, Nur77-eGFP, to demonstrate that although BCR signaling is reduced among GC B cells, a small population of cells exhibiting GC light zone phenotype (site of Ag and follicular helper T cell encounter) express much higher levels of GFP. We show that these cells exhibit somatic hypermutation, gene expression characteristic of signaling and selection, and undergo BCR signaling in vivo.
Copyright © 2015 by The American Association of Immunologists, Inc.
Figures


References
-
- Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunol Rev. 2012;247:52–63. - PubMed
-
- Tarlinton DM. Evolution in miniature: selection, survival and distribution of antigen reactive cells in the germinal centre. Immunology and Cell Biology. 2008;86:133–138. - PubMed
-
- Victora GD, Nussenzweig MC. Germinal centers. Annual Review of Immunology. 2012;30:429–457. - PubMed
-
- Chan TD, Brink R. Affinity-based selection and the germinal center response. Immunol Rev. 2012;247:11–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

