Coronavirus envelope (E) protein remains at the site of assembly
- PMID: 25726972
- PMCID: PMC4550588
- DOI: 10.1016/j.virol.2015.02.005
Coronavirus envelope (E) protein remains at the site of assembly
Abstract
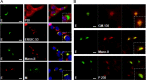
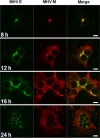
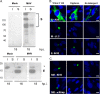
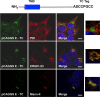
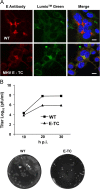
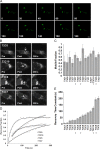
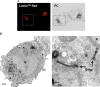
Coronaviruses (CoVs) assemble at endoplasmic reticulum Golgi intermediate compartment (ERGIC) membranes and egress from cells in cargo vesicles. Only a few molecules of the envelope (E) protein are assembled into virions. The role of E in morphogenesis is not fully understood. The cellular localization and dynamics of mouse hepatitis CoV A59 (MHV) E protein were investigated to further understanding of its role during infection. E protein localized in the ERGIC and Golgi with the amino and carboxy termini in the lumen and cytoplasm, respectively. E protein does not traffic to the cell surface. MHV was genetically engineered with a tetracysteine tag at the carboxy end of E. Fluorescence recovery after photobleaching (FRAP) showed that E is mobile in ERGIC/Golgi membranes. Correlative light electron microscopy (CLEM) confirmed the presence of E in Golgi cisternae. The results provide strong support that E proteins carry out their function(s) at the site of budding/assembly.
Keywords: CLEM; Coronavirus; Envelope protein; FRAP; Live-cell imaging; Protein transport/localization; Virus assembly.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

