Evolution of double-stranded DNA viruses of eukaryotes: from bacteriophages to transposons to giant viruses
- PMID: 25727355
- PMCID: PMC4405056
- DOI: 10.1111/nyas.12728
Evolution of double-stranded DNA viruses of eukaryotes: from bacteriophages to transposons to giant viruses
Abstract
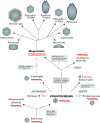
Diverse eukaryotes including animals and protists are hosts to a broad variety of viruses with double-stranded (ds) DNA genomes, from the largest known viruses, such as pandoraviruses and mimiviruses, to tiny polyomaviruses. Recent comparative genomic analyses have revealed many evolutionary connections between dsDNA viruses of eukaryotes, bacteriophages, transposable elements, and linear DNA plasmids. These findings provide an evolutionary scenario that derives several major groups of eukaryotic dsDNA viruses, including the proposed order "Megavirales," adenoviruses, and virophages from a group of large virus-like transposons known as Polintons (Mavericks). The Polintons have been recently shown to encode two capsid proteins, suggesting that these elements lead a dual lifestyle with both a transposon and a viral phase and should perhaps more appropriately be named polintoviruses. Here, we describe the recently identified evolutionary relationships between bacteriophages of the family Tectiviridae, polintoviruses, adenoviruses, virophages, large and giant DNA viruses of eukaryotes of the proposed order "Megavirales," and linear mitochondrial and cytoplasmic plasmids. We outline an evolutionary scenario under which the polintoviruses were the first group of eukaryotic dsDNA viruses that evolved from bacteriophages and became the ancestors of most large DNA viruses of eukaryotes and a variety of other selfish elements. Distinct lines of origin are detectable only for herpesviruses (from a different bacteriophage root) and polyoma/papillomaviruses (from single-stranded DNA viruses and ultimately from plasmids). Phylogenomic analysis of giant viruses provides compelling evidence of their independent origins from smaller members of the putative order "Megavirales," refuting the speculations on the evolution of these viruses from an extinct fourth domain of cellular life.
Keywords: Megavirales; Polintons; capsid proteins; translation; virus evolution.
© 2015 The Authors. Annals of the New York Academy of Sciences published by Wiley Periodicals Inc. on behalf of The New York Academy of Sciences.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

