Molecular classification of outcomes from dengue virus -3 infections
- PMID: 25728087
- PMCID: PMC4346782
- DOI: 10.1016/j.jcv.2015.01.011
Molecular classification of outcomes from dengue virus -3 infections
Abstract
Objectives: Dengue virus (DENV) infection is a significant risk to over a third of the human population that causes a wide spectrum of illness, ranging from sub-clinical disease to intermediate syndrome of vascular complications called dengue fever complicated (DFC) and severe, dengue hemorrhagic fever (DHF). Methods for discriminating outcomes will impact clinical trials and understanding disease pathophysiology.
Study design: We integrated a proteomics discovery pipeline with a heuristics approach to develop a molecular classifier to identify an intermediate phenotype of DENV-3 infectious outcome.
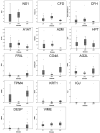
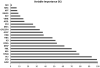
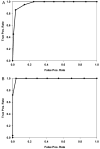
Results: 121 differentially expressed proteins were identified in plasma from DHF vs dengue fever (DF), and informative candidates were selected using nonparametric statistics. These were combined with markers that measure complement activation, acute phase response, cellular leak, granulocyte differentiation and viral load. From this, we applied quantitative proteomics to select a 15 member panel of proteins that accurately predicted DF, DHF, and DFC using a random forest classifier. The classifier primarily relied on acute phase (A2M), complement (CFD), platelet counts and cellular leak (TPM4) to produce an 86% accuracy of prediction with an area under the receiver operating curve of >0.9 for DHF and DFC vs DF.
Conclusions: Integrating discovery and heuristic approaches to sample distinct pathophysiological processes is a powerful approach in infectious disease. Early detection of intermediate outcomes of DENV-3 will speed clinical trials evaluating vaccines or drug interventions.
Keywords: Acute phase reaction; Biomarker pipeline; Dengue; Selected reaction monitoring.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures
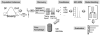



References
-
- Whitehead SS, Blaney JE, Durbin AP, Murphy BR. Prospects for a dengue virus vaccine. Nature reviews Microbiology. 2007;5(7):518–28. - PubMed
-
- Chaturvedi U, Nagar R, Shrivastava R. Dengue and dengue haemorrhagic fever: implications of host genetics. FEMS immunology and medical microbiology. 2006;47(2):155–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

