Bursty gene expression in the intact mammalian liver
- PMID: 25728770
- PMCID: PMC4500162
- DOI: 10.1016/j.molcel.2015.01.027
Bursty gene expression in the intact mammalian liver
Abstract
Bursts of nascent mRNA have been shown to lead to substantial cell-cell variation in unicellular organisms, facilitating diverse responses to environmental challenges. It is unknown whether similar bursts and gene-expression noise occur in mammalian tissues. To address this, we combine single molecule transcript counting with dual-color labeling and quantification of nascent mRNA to characterize promoter states, transcription rates, and transcript lifetimes in the intact mouse liver. We find that liver gene expression is highly bursty, with promoters stochastically switching between transcriptionally active and inactive states. Promoters of genes with short mRNA lifetimes are active longer, facilitating rapid response while reducing burst-associated noise. Moreover, polyploid hepatocytes exhibit less noise than diploid hepatocytes, suggesting a possible benefit to liver polyploidy. Thus, temporal averaging and liver polyploidy dampen the intrinsic variability associated with transcriptional bursts. Our approach can be used to study transcriptional bursting in diverse mammalian tissues.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
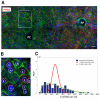
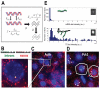
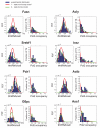
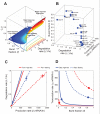


Comment in
-
A noisy tug of war: the battle between transcript production and degradation in the liver.Dev Cell. 2015 Apr 6;33(1):3-4. doi: 10.1016/j.devcel.2015.03.019. Dev Cell. 2015. PMID: 25850670
References
-
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dollé MET, Calder RB, Chisholm GB, Pollock BH, Klein CA, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. - PubMed
-
- Blake WJ, KÆrn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. - PubMed
-
- Blake WJ, Balázsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Phenotypic Consequences of Promoter-Mediated Transcriptional Noise. Mol. Cell. 2006;24:853–865. - PubMed
-
- Braeuning A, Ittrich C, Köhle C, Hailfinger S, Bonin M, Buchmann A, Schwarz M. Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 2006;273:5051–5061. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

