Modular organization of the white spruce (Picea glauca) transcriptome reveals functional organization and evolutionary signatures
- PMID: 25728802
- PMCID: PMC5024012
- DOI: 10.1111/nph.13343
Modular organization of the white spruce (Picea glauca) transcriptome reveals functional organization and evolutionary signatures
Abstract
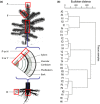
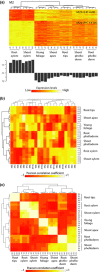
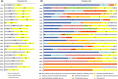
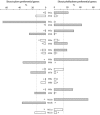
Transcript profiling has shown the molecular bases of several biological processes in plants but few studies have developed an understanding of overall transcriptome variation. We investigated transcriptome structure in white spruce (Picea glauca), aiming to delineate its modular organization and associated functional and evolutionary attributes. Microarray analyses were used to: identify and functionally characterize groups of co-expressed genes; investigate expressional and functional diversity of vascular tissue preferential genes which were conserved among Picea species, and identify expression networks underlying wood formation. We classified 22 857 genes as variable (79%; 22 coexpression groups) or invariant (21%) by profiling across several vegetative tissues. Modular organization and complex transcriptome restructuring among vascular tissue preferential genes was revealed by their assignment to coexpression groups with partially overlapping profiles and partially distinct functions. Integrated analyses of tissue-based and temporally variable profiles identified secondary xylem gene networks, showed their remodelling over a growing season and identified PgNAC-7 (no apical meristerm (NAM), Arabidopsis transcription activation factor (ATAF) and cup-shaped cotyledon (CUC) transcription factor 007 in Picea glauca) as a major hub gene specific to earlywood formation. Reference profiling identified comprehensive, statistically robust coexpressed groups, revealing that modular organization underpins the evolutionary conservation of the transcriptome structure.
Keywords: conifers; modular organization; tissue differentiation; transcriptional network; transcriptome; white spruce (Picea glauca).
© 2015 The Authors. New Phytologist © 2015 New Phytologist Trust.
Figures

References
-
- Arabidopsis Genome Initiative . 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature 408: 796. - PubMed
-
- Baldi P, Pedron L, Hietala AM, La Porta N. 2011. Cold tolerance in cypress (Cupressus sempervirens L.): a physiological and molecular study. Tree Genetics & Genomes 7: 79–90.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

