IL-7 coordinates proliferation, differentiation and Tcra recombination during thymocyte β-selection
- PMID: 25729925
- PMCID: PMC4368453
- DOI: 10.1038/ni.3122
IL-7 coordinates proliferation, differentiation and Tcra recombination during thymocyte β-selection
Abstract
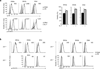
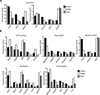
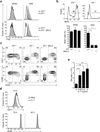
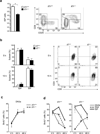
Signaling via the pre-T cell antigen receptor (pre-TCR) and the receptor Notch1 induces transient self-renewal (β-selection) of TCRβ(+) CD4(-)CD8(-) double-negative stage 3 (DN3) and DN4 progenitor cells that differentiate into CD4(+)CD8(+) double-positive (DP) thymocytes, which then rearrange the locus encoding the TCR α-chain (Tcra). Interleukin 7 (IL-7) promotes the survival of TCRβ(-) DN thymocytes by inducing expression of the pro-survival molecule Bcl-2, but the functions of IL-7 during β-selection have remained unclear. Here we found that IL-7 signaled TCRβ(+) DN3 and DN4 thymocytes to upregulate genes encoding molecules involved in cell growth and repressed the gene encoding the transcriptional repressor Bcl-6. Accordingly, IL-7-deficient DN4 cells lacked trophic receptors and did not proliferate but rearranged Tcra prematurely and differentiated rapidly. Deletion of Bcl6 partially restored the self-renewal of DN4 cells in the absence of IL-7, but overexpression of BCL2 did not. Thus, IL-7 critically acts cooperatively with signaling via the pre-TCR and Notch1 to coordinate proliferation, differentiation and Tcra recombination during β-selection.
Figures




Comment in
-
Thymic IL-7 signaling goes beyond survival.Nat Immunol. 2015 Apr;16(4):337-8. doi: 10.1038/ni.3128. Nat Immunol. 2015. PMID: 25789675 No abstract available.
References
-
- Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ. Functions of notch signaling in the immune system: consensus and controversies. Annual review of immunology. 2010;28:343–365. - PubMed
-
- Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: intelligent design. Nature reviews Immunology. 2007;7(2):144–154. - PubMed
-
- Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman IL. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 1997;89(7):1033–1041. - PubMed
-
- Maraskovsky E, O’Reilly LA, Teepe M, Corcoran LM, Peschon JJ, Strasser A. Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor-deficient mice but not in mutant rag-1−/− mice. Cell. 1997;89(7):1011–1019. - PubMed
-
- Khaled AR, Li WQ, Huang J, Fry TJ, Khaled AS, Mackall CL, et al. Bax deficiency partially corrects interleukin-7 receptor alpha deficiency. Immunity. 2002;17(5):561–573. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

