Viremic control and viral coreceptor usage in two HIV-1-infected persons homozygous for CCR5 Δ32
- PMID: 25730507
- PMCID: PMC4473772
- DOI: 10.1097/QAD.0000000000000629
Viremic control and viral coreceptor usage in two HIV-1-infected persons homozygous for CCR5 Δ32
Abstract
Objectives: To determine viral and immune factors involved in transmission and control of HIV-1 infection in persons without functional CCR5.
Design: Understanding transmission and control of HIV-1 in persons homozygous for CCR5(Δ32) is important given efforts to develop HIV-1 curative therapies aimed at modifying or disrupting CCR5 expression.
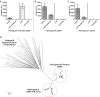
Methods: We identified two HIV-infected CCR5(Δ32/Δ32) individuals among a cohort of patients with spontaneous control of HIV-1 infection without antiretroviral therapy and determined coreceptor usage of the infecting viruses. We assessed genetic evolution of full-length HIV-1 envelope sequences by single-genome analysis from one participant and his sexual partner, and explored HIV-1 immune responses and HIV-1 mutations following virologic escape and disease progression.
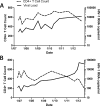
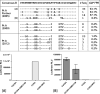
Results: Both participants experienced viremia of less than 4000 RNA copies/ml with preserved CD4(+) T-cell counts off antiretroviral therapy for at least 3.3 and 4.6 years after diagnosis, respectively. One participant had phenotypic evidence of X4 virus, had no known favorable human leukocyte antigen alleles, and appeared to be infected by minority X4 virus from a pool that predominately used CCR5 for entry. The second participant had virus that was unable to use CXCR4 for entry in phenotypic assay but was able to engage alternative viral coreceptors (e.g., CXCR6) in vitro.
Conclusion: Our study demonstrates that individuals may be infected by minority X4 viruses from a population that predominately uses CCR5 for entry, and that viruses may bypass traditional HIV-1 coreceptors (CCR5 and CXCR4) completely by engaging alternative coreceptors to establish and propagate HIV-1 infection.
Conflict of interest statement
Figures

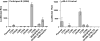
References
-
- Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis. 2008;197:563–571. - PubMed
-
- Balotta C, Bagnarelli P, Violin M, Ridolfo AL, Zhou D, Berlusconi A, et al. Homozygous delta 32 deletion of the CCR-5 chemokine receptor gene in an HIV-1-infected patient. Aids. 1997;11:F67–71. - PubMed
-
- O’Brien TR, Winkler C, Dean M, Nelson JA, Carrington M, Michael NL, et al. HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet. 1997;349:1219. - PubMed

