Whole genome sequencing in clinical and public health microbiology
- PMID: 25730631
- PMCID: PMC4389090
- DOI: 10.1097/PAT.0000000000000235
Whole genome sequencing in clinical and public health microbiology
Abstract
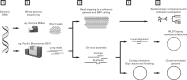
Genomics and whole genome sequencing (WGS) have the capacity to greatly enhance knowledge and understanding of infectious diseases and clinical microbiology.The growth and availability of bench-top WGS analysers has facilitated the feasibility of genomics in clinical and public health microbiology.Given current resource and infrastructure limitations, WGS is most applicable to use in public health laboratories, reference laboratories, and hospital infection control-affiliated laboratories.As WGS represents the pinnacle for strain characterisation and epidemiological analyses, it is likely to replace traditional typing methods, resistance gene detection and other sequence-based investigations (e.g., 16S rDNA PCR) in the near future.Although genomic technologies are rapidly evolving, widespread implementation in clinical and public health microbiology laboratories is limited by the need for effective semi-automated pipelines, standardised quality control and data interpretation, bioinformatics expertise, and infrastructure.
Figures

References
-
- Kuska B. Beer, Bethesda, and biology: how “genomics” came into being. J Natl Cancer Inst 1998; 90:93. - PubMed
-
- Green ED. Strategies for the systematic sequencing of complex genomes. Nat Rev Genet 2001; 2:573–583. - PubMed
-
- Fleischmann RD, Adams MD, White O, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 1995; 269:496–512. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
