Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue
- PMID: 25730763
- PMCID: PMC4380509
- DOI: 10.1038/ng.3221
Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue
Erratum in
-
Corrigendum: analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue.Nat Genet. 2015 Jun;47(6):689. doi: 10.1038/ng0615-689b. Nat Genet. 2015. PMID: 26018901 No abstract available.
Abstract
Genome-wide DNA sequencing was used to decrypt the phylogeny of multiple samples from distinct areas of cancer and morphologically normal tissue taken from the prostates of three men. Mutations were present at high levels in morphologically normal tissue distant from the cancer, reflecting clonal expansions, and the underlying mutational processes at work in morphologically normal tissue were also at work in cancer. Our observations demonstrate the existence of ongoing abnormal mutational processes, consistent with field effects, underlying carcinogenesis. This mechanism gives rise to extensive branching evolution and cancer clone mixing, as exemplified by the coexistence of multiple cancer lineages harboring distinct ERG fusions within a single cancer nodule. Subsets of mutations were shared either by morphologically normal and malignant tissues or between different ERG lineages, indicating earlier or separate clonal cell expansions. Our observations inform on the origin of multifocal disease and have implications for prostate cancer therapy in individual cases.
Figures
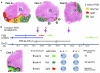
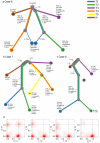

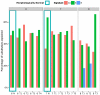
Comment in
-
Prostate cancer: Multifocal disease--independent clonal lineages in malignant nodules and even in normal tissue.Nat Rev Urol. 2015 Apr;12(4):177. doi: 10.1038/nrurol.2015.52. Epub 2015 Mar 17. Nat Rev Urol. 2015. PMID: 25782174 No abstract available.
References
REFERENCE LIST
-
- Andreoiu M, Cheng L. Multifocal prostate cancer: biologic, prognostic, and therapeutic implications. Hum. Pathol. 2010;41:781–793. - PubMed
-
- Cheng L, et al. Evidence of independent origin of multiple tumors from patients with prostate cancer. J. Natl. Cancer Inst. 1998;90:233–237. - PubMed
-
- Kobayashi M, et al. Molecular analysis of multifocal prostate cancer by comparative genomic hybridization. Prostate. 2008;68:1715–1724. - PubMed
-
- Boyd LK, et al. High-resolution genome-wide copy-number analysis suggests a monoclonal origin of multifocal prostate cancer. Genes Chromosomes Cancer. 2012;51:579–589. - PubMed
-
- Lindberg J, et al. Exome sequencing of prostate cancer supports the hypothesis of independent tumour origins. Eur. Urol. 2013;63:347–353. - PubMed
METHODS-ONLY REFERENCES
-
- Warren AY, et al. Method for sampling tissue for research which preserves pathological data in radical prostatectomy. Prostate. 2013;73:194–202. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

