Notum deacylates Wnt proteins to suppress signalling activity
- PMID: 25731175
- PMCID: PMC4376489
- DOI: 10.1038/nature14259
Notum deacylates Wnt proteins to suppress signalling activity
Abstract
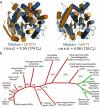
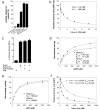
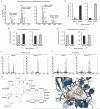
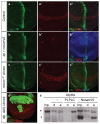
Signalling by Wnt proteins is finely balanced to ensure normal development and tissue homeostasis while avoiding diseases such as cancer. This is achieved in part by Notum, a highly conserved secreted feedback antagonist. Notum has been thought to act as a phospholipase, shedding glypicans and associated Wnt proteins from the cell surface. However, this view fails to explain specificity, as glypicans bind many extracellular ligands. Here we provide genetic evidence in Drosophila that Notum requires glypicans to suppress Wnt signalling, but does not cleave their glycophosphatidylinositol anchor. Structural analyses reveal glycosaminoglycan binding sites on Notum, which probably help Notum to co-localize with Wnt proteins. They also identify, at the active site of human and Drosophila Notum, a large hydrophobic pocket that accommodates palmitoleate. Kinetic and mass spectrometric analyses of human proteins show that Notum is a carboxylesterase that removes an essential palmitoleate moiety from Wnt proteins and thus constitutes the first known extracellular protein deacylase.
Figures
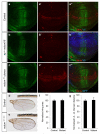
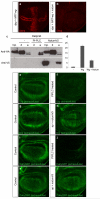
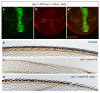
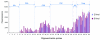


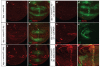

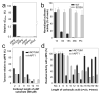
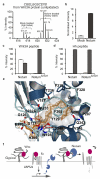
Comment in
-
Cell signalling: Disarming Wnt.Nature. 2015 Mar 12;519(7542):163-4. doi: 10.1038/nature14208. Epub 2015 Feb 25. Nature. 2015. PMID: 25731170 No abstract available.
-
Development: Switching off WNT with precision.Nat Rev Mol Cell Biol. 2015 Apr;16(4):204. doi: 10.1038/nrm3971. Epub 2015 Mar 18. Nat Rev Mol Cell Biol. 2015. PMID: 25785717 No abstract available.
References
-
- Freeman M. Feedback control of intercellular signalling in development. Nature. 2000;408:313–319. - PubMed
-
- Takada R, et al. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev. Cell. 2006;11:791–801. - PubMed
-
- Willert K, et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. - PubMed
Methods references
Extended data reference
-
- Glise B, et al. Shifted, the Drosophila ortholog of Wnt inhibitory factor-1, controls the distribution and movement of Hedgehog. Dev. Cell. 2005;8:255–266. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- 17721/CRUK_/Cancer Research UK/United Kingdom
- WT099197MA/WT_/Wellcome Trust/United Kingdom
- G0900084/MRC_/Medical Research Council/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- 090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- 294523/ERC_/European Research Council/International
- C375/A10976/CRUK_/Cancer Research UK/United Kingdom
- MC_U117584268/MRC_/Medical Research Council/United Kingdom
- A10976/CRUK_/Cancer Research UK/United Kingdom
- WT093378MA/WT_/Wellcome Trust/United Kingdom
- U117584268/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

